Fragmentation landscape of cell-free DNA revealed by deconvolutional analysis of end motifs
- PMID: 37075072
- PMCID: PMC10151549
- DOI: 10.1073/pnas.2220982120
Fragmentation landscape of cell-free DNA revealed by deconvolutional analysis of end motifs
Abstract
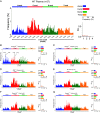
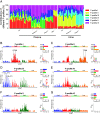
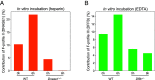
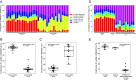
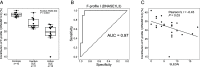
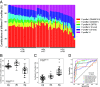
Cell-free DNA (cfDNA) fragmentation is nonrandom, at least partially mediated by various DNA nucleases, forming characteristic cfDNA end motifs. However, there is a paucity of tools for deciphering the relative contributions of cfDNA cleavage patterns related to underlying fragmentation factors. In this study, through non-negative matrix factorization algorithm, we used 256 5' 4-mer end motifs to identify distinct types of cfDNA cleavage patterns, referred to as "founder" end-motif profiles (F-profiles). F-profiles were associated with different DNA nucleases based on whether such patterns were disrupted in nuclease-knockout mouse models. Contributions of individual F-profiles in a cfDNA sample could be determined by deconvolutional analysis. We analyzed 93 murine cfDNA samples of different nuclease-deficient mice and identified six types of F-profiles. F-profiles I, II, and III were linked to deoxyribonuclease 1 like 3 (DNASE1L3), deoxyribonuclease 1 (DNASE1), and DNA fragmentation factor subunit beta (DFFB), respectively. We revealed that 42.9% of plasma cfDNA molecules were attributed to DNASE1L3-mediated fragmentation, whereas 43.4% of urinary cfDNA molecules involved DNASE1-mediated fragmentation. We further demonstrated that the relative contributions of F-profiles were useful to inform pathological states, such as autoimmune disorders and cancer. Among the six F-profiles, the use of F-profile I could inform the human patients with systemic lupus erythematosus. F-profile VI could be used to detect individuals with hepatocellular carcinoma, with an area under the receiver operating characteristic curve of 0.97. F-profile VI was more prominent in patients with nasopharyngeal carcinoma undergoing chemoradiotherapy. We proposed that this profile might be related to oxidative stress.
Keywords: cancer detection; fragmentomics; liquid biopsy; non-negative matrix factorization; oxidative stress.
Conflict of interest statement
The authors have organizational affiliations to disclose, K.C.A.C. and Y.M.D.L. hold leadership positions in Centre for Novostics. R.W.K.C., K.C.A.C., and Y.M.D.L. hold equities in Take2. Z.Z., M.-J.L.M., K.C.A.C., Y.M.D.L., and P.J. have filed a patent application on the described technology.
Figures


References
-
- Lo Y. M. D., et al. , Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2, 61ra91 (2010). - PubMed
-
- Lo Y. M. D., Han D. S. C., Jiang P., Chiu R. W. K., Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 372, eaaw3616 (2021). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

