A rarefaction approach for measuring population differences in rare and common variation
- PMID: 37075098
- PMCID: PMC10213490
- DOI: 10.1093/genetics/iyad070
A rarefaction approach for measuring population differences in rare and common variation
Abstract
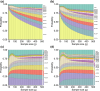
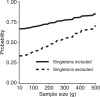
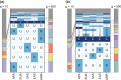
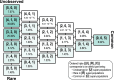
In studying allele-frequency variation across populations, it is often convenient to classify an allelic type as "rare," with nonzero frequency less than or equal to a specified threshold, "common," with a frequency above the threshold, or entirely unobserved in a population. When sample sizes differ across populations, however, especially if the threshold separating "rare" and "common" corresponds to a small number of observed copies of an allelic type, discreteness effects can lead a sample from one population to possess substantially more rare allelic types than a sample from another population, even if the two populations have extremely similar underlying allele-frequency distributions across loci. We introduce a rarefaction-based sample-size correction for use in comparing rare and common variation across multiple populations whose sample sizes potentially differ. We use our approach to examine rare and common variation in worldwide human populations, finding that the sample-size correction introduces subtle differences relative to analyses that use the full available sample sizes. We introduce several ways in which the rarefaction approach can be applied: we explore the dependence of allele classifications on subsample sizes, we permit more than two classes of allelic types of nonzero frequency, and we analyze rare and common variation in sliding windows along the genome. The results can assist in clarifying similarities and differences in allele-frequency patterns across populations.
Keywords: common variants; rare variants; rarefaction; sample-size correction.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflict of interest.
Figures
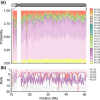
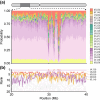
References
-
- Byrska-Bishop M, Evani US, Zhao X, Basile AO, Abel HJ, Regier AA, Corvelo A, Clarke WE, Musunuri R, Nagulapalli K, et al. High-coverage whole-genome sequencing of the expanded 1000 genomes project cohort including 602 trios. Cell. 2022;185:3426–3440.e19. doi:10.1016/j.cell.2022.08.004 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

