PI3Kβ controls immune evasion in PTEN-deficient breast tumours
- PMID: 37076617
- PMCID: PMC10494520
- DOI: 10.1038/s41586-023-05940-w
PI3Kβ controls immune evasion in PTEN-deficient breast tumours
Abstract
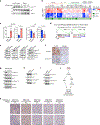
Loss of the PTEN tumour suppressor is one of the most common oncogenic drivers across all cancer types1. PTEN is the major negative regulator of PI3K signalling. The PI3Kβ isoform has been shown to play an important role in PTEN-deficient tumours, but the mechanisms underlying the importance of PI3Kβ activity remain elusive. Here, using a syngeneic genetically engineered mouse model of invasive breast cancer driven by ablation of both Pten and Trp53 (which encodes p53), we show that genetic inactivation of PI3Kβ led to a robust anti-tumour immune response that abrogated tumour growth in syngeneic immunocompetent mice, but not in immunodeficient mice. Mechanistically, PI3Kβ inactivation in the PTEN-null setting led to reduced STAT3 signalling and increased the expression of immune stimulatory molecules, thereby promoting anti-tumour immune responses. Pharmacological PI3Kβ inhibition also elicited anti-tumour immunity and synergized with immunotherapy to inhibit tumour growth. Mice with complete responses to the combined treatment displayed immune memory and rejected tumours upon re-challenge. Our findings demonstrate a molecular mechanism linking PTEN loss and STAT3 activation in cancer and suggest that PI3Kβ controls immune escape in PTEN-null tumours, providing a rationale for combining PI3Kβ inhibitors with immunotherapy for the treatment of PTEN-deficient breast cancer.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
J.S.B. is a scientific consultant for Geode Therapeutics Inc. J.S.B., G.J.F., T.M.R. and J.J.Z. are co-inventors of DFCI 2180.001 (DFS-166.25) related to this work. Qiwei W. is a scientific consultant for Crimson Biopharm Inc. Qi W. is currently an employee at Geode Therapeutics Inc. S.P. is currently an employee at Takeda Pharmaceuticals. S.X. is currently an employee at HiFiBiO Therapeutics. T.V. is currently an employee at MarvelBiome Inc. G.J.F. has patents/pending royalties on the PD-1/PD-L1 pathway from Roche, Merck MSD, Bristol-Myers-Squibb, Merck KGA, Boehringer-Ingelheim, AstraZeneca, Dako, Leica, Mayo Clinic, Eli Lilly and Novartis. G.J.F. has served on advisory boards for Roche, Bristol-Myers-Squibb, Xios, Origimed, Triursus, iTeos, NextPoint, IgM, Jubilant, Trillium, IOME, Geode, Bright Peak, and GV20. G.J.F. has equity in Nextpoint, Triursus, Xios, iTeos, IgM, Trillium, Invaria, Geode, and GV20. H-J.K. is currently an employee at Genentech. A.K.S. has received compensation for consulting and/or SAB membership from Merck, Honeycomb Biotechnologies, Cellarity, Repertoire Immune Medicines, Hovione, Third Rock Ventures, Ochre Bio, FL82, Empress Therapeutics, Relation Therapeutics, Senda Biosciences, IntrECate biotherapeutics, Santa Ana Bio, and Dahlia Biosciences unrelated to this work. T.M.R. is a SAB member for Shiftbio and K2B Therapeutics and is a co-founder of Geode Therapeutics Inc. J.J.Z. is a co-founder and board director of Crimson Biotech Inc. and Geode Therapeutics Inc. All other authors declare no competing interests.
Figures










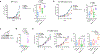
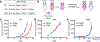


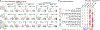
Comment in
-
Enzyme lights dual fires to promote cancer.Nature. 2023 May;617(7959):42-43. doi: 10.1038/d41586-023-01025-w. Nature. 2023. PMID: 37076711 No abstract available.
References
Additional references
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

