Dedifferentiation maintains melanocyte stem cells in a dynamic niche
- PMID: 37076619
- PMCID: PMC10132989
- DOI: 10.1038/s41586-023-05960-6
Dedifferentiation maintains melanocyte stem cells in a dynamic niche
Abstract
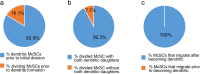
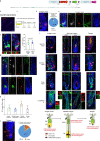
For unknow reasons, the melanocyte stem cell (McSC) system fails earlier than other adult stem cell populations1, which leads to hair greying in most humans and mice2,3. Current dogma states that McSCs are reserved in an undifferentiated state in the hair follicle niche, physically segregated from differentiated progeny that migrate away following cues of regenerative stimuli4-8. Here we show that most McSCs toggle between transit-amplifying and stem cell states for both self-renewal and generation of mature progeny, a mechanism fundamentally distinct from those of other self-renewing systems. Live imaging and single-cell RNA sequencing revealed that McSCs are mobile, translocating between hair follicle stem cell and transit-amplifying compartments where they reversibly enter distinct differentiation states governed by local microenvironmental cues (for example, WNT). Long-term lineage tracing demonstrated that the McSC system is maintained by reverted McSCs rather than by reserved stem cells inherently exempt from reversible changes. During ageing, there is accumulation of stranded McSCs that do not contribute to the regeneration of melanocyte progeny. These results identify a new model whereby dedifferentiation is integral to homeostatic stem cell maintenance and suggest that modulating McSC mobility may represent a new approach for the prevention of hair greying.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures















Comment in
-
Yo-yoing stem cells defy dogma to maintain hair colour.Nature. 2023 Apr;616(7958):666-667. doi: 10.1038/d41586-023-00918-0. Nature. 2023. PMID: 37076710 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

