Reintroducing genetic diversity in populations from cryopreserved material: the case of Abondance, a French local dairy cattle breed
- PMID: 37076793
- PMCID: PMC10114384
- DOI: 10.1186/s12711-023-00801-6
Reintroducing genetic diversity in populations from cryopreserved material: the case of Abondance, a French local dairy cattle breed
Abstract
Background: Genetic diversity is a necessary condition for populations to evolve under natural adaptation, artificial selection, or both. However, genetic diversity is often threatened, in particular in domestic animal populations where artificial selection, genetic drift and inbreeding are strong. In this context, cryopreserved genetic resources are a promising option to reintroduce lost variants and to limit inbreeding. However, while the use of ancient genetic resources is more common in plant breeding, it is less documented in animals due to a longer generation interval, making it difficult to fill the gap in performance due to continuous selection. This study investigates one of the only concrete cases available in animals, for which cryopreserved semen from a bull born in 1977 in a lost lineage was introduced into the breeding scheme of a French local dairy cattle breed, the Abondance breed, more than 20 years later.
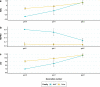
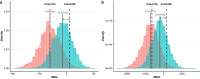
Results: We found that this re-introduced bull was genetically distinct with respect to the current population and thus allowed part of the genetic diversity lost over time to be restored. The expected negative gap in milk production due to continuous selection was absorbed in a few years by preferential mating with elite cows. Moreover, the re-use of this bull more than two decades later did not increase the level of inbreeding, and even tended to reduce it by avoiding mating with relatives. Finally, the reintroduction of a bull from a lost lineage in the breeding scheme allowed for improved performance for reproductive abilities, a trait that was less subject to selection in the past.
Conclusions: The use of cryopreserved material is an efficient way to manage the genetic diversity of an animal population, by mitigating the effects of both inbreeding and strong selection. However, attention should be paid to mating of animals to limit the disadvantages associated with incorporating original genetic material, notably a discrepancy in the breeding values for selected traits or an increase in inbreeding. Therefore, careful characterization of the genetic resources available in cryobanks could help to ensure the sustainable management of populations, in particular local or small populations. These results could also be transferred to the conservation of wild threatened populations.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

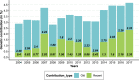

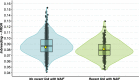
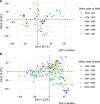
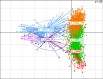

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

