Genomic insight of sulfate reducing bacterial genus Desulfofaba reveals their metabolic versatility in biogeochemical cycling
- PMID: 37076818
- PMCID: PMC10116758
- DOI: 10.1186/s12864-023-09297-2
Genomic insight of sulfate reducing bacterial genus Desulfofaba reveals their metabolic versatility in biogeochemical cycling
Abstract
Background: Sulfate-reducing bacteria (SRB) drive the ocean sulfur and carbon cycling. They constitute a diverse phylogenetic and physiological group and are widely distributed in anoxic marine environments. From a physiological viewpoint, SRB's can be categorized as complete or incomplete oxidizers, meaning that they either oxidize their carbon substrate completely to CO2 or to a stoichiometric mix of CO2 and acetate. Members of Desulfofabaceae family are incomplete oxidizers, and within that family, Desulfofaba is the only genus with three isolates that are classified into three species. Previous physiological experiments revealed their capability of respiring oxygen.
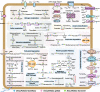
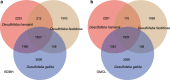
Results: Here, we sequenced the genomes of three isolates in Desulfofaba genus and reported on a genomic comparison of the three species to reveal their metabolic potentials. Based on their genomic contents, they all could oxidize propionate to acetate and CO2. We confirmed their phylogenetic position as incomplete oxidizers based on dissimilatory sulfate reductase (DsrAB) phylogeny. We found the complete pathway for dissimilatory sulfate reduction, but also different key genes for nitrogen cycling, including nitrogen fixation, assimilatory nitrate/nitrite reduction, and hydroxylamine reduction to nitrous oxide. Their genomes also contain genes that allow them to cope with oxygen and oxidative stress. They have genes that encode for diverse central metabolisms for utilizing different substrates with the potential for more strains to be isolated in the future, yet their distribution is limited.
Conclusions: Results based on marker gene search and curated metagenome assembled genomes search suggest a limited environmental distribution of this genus. Our results reveal a large metabolic versatility within the Desulfofaba genus which establishes their importance in biogeochemical cycling of carbon in their respective habitats, as well as in the support of the entire microbial community through releasing easily degraded organic matters.
Keywords: Desulfofaba; Genome; Incomplete sulfate reducer; Nitrogen; Oxygen; Sulfur.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Rice Paddy Nitrospirae Carry and Express Genes Related to Sulfate Respiration: Proposal of the New Genus "Candidatus Sulfobium".Appl Environ Microbiol. 2018 Feb 14;84(5):e02224-17. doi: 10.1128/AEM.02224-17. Print 2018 Mar 1. Appl Environ Microbiol. 2018. PMID: 29247059 Free PMC article.
-
Characterization of the marine propionate-degrading, sulfate-reducing bacterium Desulfofaba fastidiosa sp. nov. and reclassification of Desulfomusa hansenii as Desulfofaba hansenii comb. nov.Int J Syst Evol Microbiol. 2004 Mar;54(Pt 2):393-399. doi: 10.1099/ijs.0.02820-0. Int J Syst Evol Microbiol. 2004. PMID: 15023950
-
Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes.J Bacteriol. 2001 Oct;183(20):6028-35. doi: 10.1128/JB.183.20.6028-6035.2001. J Bacteriol. 2001. PMID: 11567003 Free PMC article.
-
Dissimilatory sulfur cycling in oxygen minimum zones: an emerging metagenomics perspective.Biochem Soc Trans. 2011 Dec;39(6):1859-63. doi: 10.1042/BST20110708. Biochem Soc Trans. 2011. PMID: 22103540 Review.
-
The role of sulfate-reducing prokaryotes in the coupling of element biogeochemical cycling.Sci Total Environ. 2018 Feb 1;613-614:398-408. doi: 10.1016/j.scitotenv.2017.09.062. Epub 2017 Sep 26. Sci Total Environ. 2018. PMID: 28918271 Review.
Cited by
-
Unveiling the vital role of soil microorganisms in selenium cycling: a review.Front Microbiol. 2024 Sep 11;15:1448539. doi: 10.3389/fmicb.2024.1448539. eCollection 2024. Front Microbiol. 2024. PMID: 39323878 Free PMC article. Review.
-
Chaihushugan powder regulates the gut microbiota to alleviate mitochondrial oxidative stress in the gastric tissues of rats with functional dyspepsia.Front Immunol. 2025 Feb 18;16:1549554. doi: 10.3389/fimmu.2025.1549554. eCollection 2025. Front Immunol. 2025. PMID: 40040709 Free PMC article.
-
Sulfate-reducing bacteria unearthed: ecological functions of the diverse prokaryotic group in terrestrial environments.Appl Environ Microbiol. 2024 Apr 17;90(4):e0139023. doi: 10.1128/aem.01390-23. Epub 2024 Mar 29. Appl Environ Microbiol. 2024. PMID: 38551370 Free PMC article. Review.
-
Virtual screening and molecular dynamic simulations of Desulfovibrio vulgaris dissimilatory sulfite reductase inhibitors for the control of sulphate reducing bacteria.In Silico Pharmacol. 2025 Jun 17;13(2):90. doi: 10.1007/s40203-025-00367-9. eCollection 2025. In Silico Pharmacol. 2025. PMID: 40539084
-
Influence of Mikania micrantha Kunth Flavonoids on Composition of Soil Microbial Community.Int J Mol Sci. 2024 Dec 25;26(1):64. doi: 10.3390/ijms26010064. Int J Mol Sci. 2024. PMID: 39795923 Free PMC article.
References
-
- Jørgensen BB. The sulfur cycle of freshwater sediments: Role of thiosulfate. Limnol Oceanogr. 1990;35:1329–1342. doi: 10.4319/lo.1990.35.6.1329. - DOI
-
- Jørgensen BB. Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature. 1982;296:643–645. doi: 10.1038/296643a0. - DOI
-
- Cypionka H, Widdel F, Pfennig N. Survival of sulfate-reducing bacteria after oxygen stress, and growth in sulfate-free oxygen-sulfide gradients. FEMS Microbiol Ecol. 1985;1:39–45. doi: 10.1111/j.1574-6968.1985.tb01129.x. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

