High curvature promotes fusion of lipid membranes: Predictions from continuum elastic theory
- PMID: 37077047
- PMCID: PMC10209146
- DOI: 10.1016/j.bpj.2023.04.018
High curvature promotes fusion of lipid membranes: Predictions from continuum elastic theory
Abstract
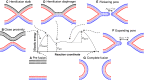
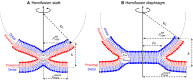
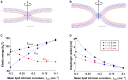
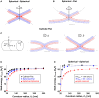
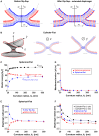
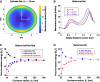
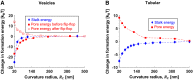
The fusion of lipid membranes progresses through a series of hemifusion intermediates with two significant energy barriers related to the formation of stalk and fusion pore, respectively. These energy barriers determine the speed and success rate of many critical biological processes, including the fusion of highly curved membranes, for example synaptic vesicles and enveloped viruses. Here we use continuum elastic theory of lipid monolayers to determine the relationship between membrane shape and energy barriers to fusion. We find that the stalk formation energy decreases with curvature by up to 31 kBT in a 20-nm-radius vesicle compared with planar membranes and by up to 8 kBT in the fusion of highly curved, long, tubular membranes. In contrast, the fusion pore formation energy barrier shows a more complicated behavior. Immediately after stalk expansion to the hemifusion diaphragm, the fusion pore formation energy barrier is low (15-25 kBT) due to lipid stretching in the distal monolayers and increased tension in highly curved vesicles. Therefore, the opening of the fusion pore is faster. However, these stresses relax over time due to lipid flip-flop from the proximal monolayer, resulting in a larger hemifusion diaphragm and a higher fusion pore formation energy barrier, up to 35 kBT. Therefore, if the fusion pore fails to open before significant lipid flip-flop takes place, the reaction proceeds to an extended hemifusion diaphragm state, which is a dead-end configuration in the fusion process and can be used to prevent viral infections. In contrast, in the fusion of long tubular compartments, the surface tension does not accumulate due to the formation of the diaphragm, and the energy barrier for pore expansion increases with curvature by up to 11 kBT. This suggests that inhibition of polymorphic virus infection could particularly target this feature of the second barrier.
Copyright © 2023 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
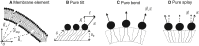
References
-
- Jahn R., Grubmüller H. Membrane fusion. Curr. Opin. Cell Biol. 2002;14:488–495. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

