Comparative transcriptomics reveals desynchronisation of gene expression during the floral transition between Arabidopsis and Brassica rapa cultivars
- PMID: 37077206
- PMCID: PMC10095958
- DOI: 10.1017/qpb.2021.6
Comparative transcriptomics reveals desynchronisation of gene expression during the floral transition between Arabidopsis and Brassica rapa cultivars
Abstract
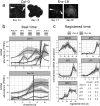
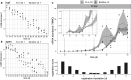
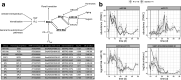
Comparative transcriptomics can be used to translate an understanding of gene regulatory networks from model systems to less studied species. Here, we use RNA-Seq to determine and compare gene expression dynamics through the floral transition in the model species Arabidopsis thaliana and the closely related crop Brassica rapa. We find that different curve registration functions are required for different genes, indicating that there is no single common 'developmental time' between Arabidopsis and B. rapa. A detailed comparison between Arabidopsis and B. rapa and between two B. rapa accessions reveals different modes of regulation of the key floral integrator SOC1, and that the floral transition in the B. rapa accessions is triggered by different pathways. Our study adds to the mechanistic understanding of the regulatory network of flowering time in rapid cycling B. rapa and highlights the importance of registration methods for the comparison of developmental gene expression data.
Keywords: Arabidopsis; Brassica rapa; FT; SOC1; comparative transcriptomics; flowering.
© The Author(s), 2021. Published by Cambridge University Press in association with The John Innes Centre 2021.
Conflict of interest statement
The authors declare no significant competing financial, professional or personal interests which might influence the performance or presentation of this study.
Figures


Similar articles
-
Brassica rapa CURLY LEAF is a major H3K27 methyltransferase regulating flowering time.Planta. 2024 Jun 12;260(1):27. doi: 10.1007/s00425-024-04454-7. Planta. 2024. PMID: 38865018 Free PMC article.
-
High ambient temperature leads to reduced FT expression and delayed flowering in Brassica rapa via a mechanism associated with H2A.Z dynamics.Plant J. 2019 Oct;100(2):343-356. doi: 10.1111/tpj.14446. Epub 2019 Aug 9. Plant J. 2019. PMID: 31257648
-
The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co-expression networks.J Exp Bot. 2013 Nov;64(14):4503-16. doi: 10.1093/jxb/ert264. Epub 2013 Sep 27. J Exp Bot. 2013. PMID: 24078668 Free PMC article.
-
Genome Triplication Leads to Transcriptional Divergence of FLOWERING LOCUS C Genes During Vernalization in the Genus Brassica.Front Plant Sci. 2021 Feb 9;11:619417. doi: 10.3389/fpls.2020.619417. eCollection 2020. Front Plant Sci. 2021. PMID: 33633752 Free PMC article. Review.
-
Gene co-expression network analysis reveals key pathways and hub genes in Chinese cabbage (Brassica rapa L.) during vernalization.BMC Genomics. 2021 Apr 6;22(1):236. doi: 10.1186/s12864-021-07510-8. BMC Genomics. 2021. PMID: 33823810 Free PMC article. Review.
Cited by
-
Flowering time: From physiology, through genetics to mechanism.Plant Physiol. 2024 Apr 30;195(1):190-212. doi: 10.1093/plphys/kiae109. Plant Physiol. 2024. PMID: 38417841 Free PMC article. Review.
-
A transcriptomic time-series reveals differing trajectories during pre-floral development in the apex and leaf in winter and spring varieties of Brassica napus.Sci Rep. 2024 Feb 12;14(1):3538. doi: 10.1038/s41598-024-53526-x. Sci Rep. 2024. PMID: 38347020 Free PMC article.
-
The 1972 Meadows report: A wake-up call for plant science.Quant Plant Biol. 2023 Mar 6;4:e3. doi: 10.1017/qpb.2023.2. eCollection 2023. Quant Plant Biol. 2023. PMID: 37077701 Free PMC article.
-
Brassica rapa CURLY LEAF is a major H3K27 methyltransferase regulating flowering time.Planta. 2024 Jun 12;260(1):27. doi: 10.1007/s00425-024-04454-7. Planta. 2024. PMID: 38865018 Free PMC article.
References
-
- Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., & Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science, 309, 1052–1056 - PubMed
-
- Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., Van Der Straeten, D., Peng, J., & Harberd, N. P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science, 311, 91–94 - PubMed
-
- Andrés, F., & Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nature Reviews Genetics, 13, 627–639 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
