The Arabidopsis embryo as a quantifiable model for studying pattern formation
- PMID: 37077211
- PMCID: PMC10095805
- DOI: 10.1017/qpb.2021.3
The Arabidopsis embryo as a quantifiable model for studying pattern formation
Abstract
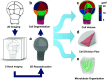
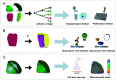
Phenotypic diversity of flowering plants stems from common basic features of the plant body pattern with well-defined body axes, organs and tissue organisation. Cell division and cell specification are the two processes that underlie the formation of a body pattern. As plant cells are encased into their cellulosic walls, directional cell division through precise positioning of division plane is crucial for shaping plant morphology. Since many plant cells are pluripotent, their fate establishment is influenced by their cellular environment through cell-to-cell signaling. Recent studies show that apart from biochemical regulation, these two processes are also influenced by cell and tissue morphology and operate under mechanical control. Finding a proper model system that allows dissecting the relationship between these aspects is the key to our understanding of pattern establishment. In this review, we present the Arabidopsis embryo as a simple, yet comprehensive model of pattern formation compatible with high-throughput quantitative assays.
Keywords: cell specification; computational cell biology; gene expression; pattern formation; plant development; plant embryogenesis.
© The Author(s) 2021.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Building a plant: cell fate specification in the early Arabidopsis embryo.Development. 2015 Feb 1;142(3):420-30. doi: 10.1242/dev.111500. Development. 2015. PMID: 25605778 Review.
-
The Arabidopsis embryo as a miniature morphogenesis model.New Phytol. 2013 Jul;199(1):14-25. doi: 10.1111/nph.12267. Epub 2013 Apr 16. New Phytol. 2013. PMID: 23590679 Review.
-
Pattern formation in plant development: four vignettes.Curr Opin Genet Dev. 1994 Aug;4(4):602-8. doi: 10.1016/0959-437x(94)90079-i. Curr Opin Genet Dev. 1994. PMID: 7950330 Review.
-
Quantitative analysis of 3D cellular geometry and modelling of the Arabidopsis embryo.J Microsc. 2022 Sep;287(3):107-113. doi: 10.1111/jmi.13130. Epub 2022 Jul 12. J Microsc. 2022. PMID: 35759505 Review.
-
Live-cell imaging and optical manipulation of Arabidopsis early embryogenesis.Dev Cell. 2015 Jul 27;34(2):242-51. doi: 10.1016/j.devcel.2015.06.008. Epub 2015 Jul 9. Dev Cell. 2015. PMID: 26166301
Cited by
-
An elastic proteinaceous envelope encapsulates the early Arabidopsis embryo.Development. 2023 Nov 15;150(22):dev201943. doi: 10.1242/dev.201943. Epub 2023 Nov 9. Development. 2023. PMID: 37869985 Free PMC article.
References
-
- Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., Nussaume, L., Noh, Y.-S., Amasino, R., & Scheres, B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell, 119, 109–120. - PubMed
-
- Barro, A. V., Stöckle, D., Thellmann, M., Ruiz-Duarte, P., Bald, L., Louveaux, M., von Born, P., Denninger, P., Goh, T., & Fukaki, H. (2019). Cytoskeleton dynamics are necessary for early events of lateral root initiation in Arabidopsis. Current Biology, 29, 2443–2454. e2445. - PubMed
-
- Bassel, G. W., Stamm, P., Mosca, G., de Reuille, P. B., Gibbs, D. J., Winter, R., Janka, A., Holdsworth, M. J., & Smith, R. S. (2014). Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proceedings of the National Academy of Sciences, 111, 8685–8690. - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
