What is quantitative plant biology?
- PMID: 37077212
- PMCID: PMC10095877
- DOI: 10.1017/qpb.2021.8
What is quantitative plant biology?
Abstract
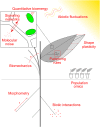
Quantitative plant biology is an interdisciplinary field that builds on a long history of biomathematics and biophysics. Today, thanks to high spatiotemporal resolution tools and computational modelling, it sets a new standard in plant science. Acquired data, whether molecular, geometric or mechanical, are quantified, statistically assessed and integrated at multiple scales and across fields. They feed testable predictions that, in turn, guide further experimental tests. Quantitative features such as variability, noise, robustness, delays or feedback loops are included to account for the inner dynamics of plants and their interactions with the environment. Here, we present the main features of this ongoing revolution, through new questions around signalling networks, tissue topology, shape plasticity, biomechanics, bioenergetics, ecology and engineering. In the end, quantitative plant biology allows us to question and better understand our interactions with plants. In turn, this field opens the door to transdisciplinary projects with the society, notably through citizen science.
© The Author(s), 2021. Published by Cambridge University Press in association with The John Innes Centre 2021.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures

References
-
- Adachi, H. , Derevnina, L. , & Kamoun, S. (2019). NLR singletons, pairs, and networks: Evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Current Opinion in Plant Biology, 50, 121–131. - PubMed
-
- Ali, O. , & Traas, J. (2016). Force-driven polymerization and turgor-Induced Wall expansion. Trends in Plant Science, 21, 398–409. - PubMed
-
- Alonso-Blanco, C. , Andrade, J. , Becker, C. , Bemm, F. , Bergelson, J. , Borgwardt, K. M. , Cao, J. , Chae, E. , Dezwaan, T. M. , Ding, W. , Ecker, J. R. , Exposito-Alonso, M. , Farlow, A. , Fitz, J. , Gan, X. , Grimm, D. G. , Hancock, A. M. , Henz, S. R. , Holm, S. , & Zhou, X. (2016). 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell, 166, 481–491. - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
