MethylScore, a pipeline for accurate and context-aware identification of differentially methylated regions from population-scale plant whole-genome bisulfite sequencing data
- PMID: 37077980
- PMCID: PMC10095865
- DOI: 10.1017/qpb.2022.14
MethylScore, a pipeline for accurate and context-aware identification of differentially methylated regions from population-scale plant whole-genome bisulfite sequencing data
Abstract
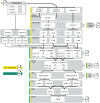
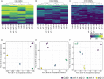
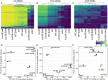
Whole-genome bisulfite sequencing (WGBS) is the standard method for profiling DNA methylation at single-nucleotide resolution. Different tools have been developed to extract differentially methylated regions (DMRs), often built upon assumptions from mammalian data. Here, we present MethylScore, a pipeline to analyse WGBS data and to account for the substantially more complex and variable nature of plant DNA methylation. MethylScore uses an unsupervised machine learning approach to segment the genome by classification into states of high and low methylation. It processes data from genomic alignments to DMR output and is designed to be usable by novice and expert users alike. We show how MethylScore can identify DMRs from hundreds of samples and how its data-driven approach can stratify associated samples without prior information. We identify DMRs in the A. thaliana 1,001 Genomes dataset to unveil known and unknown genotype-epigenotype associations .
Keywords: DNA methylation; Differential methylation; Differentially methylated regions; Epigenome; Whole-genome bisulfite sequencing; machine learning.
© The Author(s) 2022.
Conflict of interest statement
The authors declare the following competing interests: J.H. is currently an employee of Computomics GmbH. S.J.S. is currently the CEO of and holds shares in Computomics GmbH. D.W. hold shares in Computomics GmbH. A.N. is currently an employee of ecSEQ Bioinformatics GmbH. D.L. is currently the CEO of and holds shares in ecSEQ Bioinformatics GmbH.
Figures



References
-
- Becker, C. , Hagmann, J. , Müller, J. , Koenig, D. , Stegle, O. , Borgwardt, K. , & Weigel, D. (2011). Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature, 480, 245–249. - PubMed
-
- Cervera, M. T. , Ruiz-García, L. , & Martínez-Zapater, J. M. (2002). Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP markers. Molecular Genetics and Genomics, 268, 543–552. - PubMed
LinkOut - more resources
Full Text Sources
