TP53 mutations identify high-risk events for peripheral T-cell lymphoma treated with CHOP-based chemotherapy
- PMID: 37078708
- PMCID: PMC10480533
- DOI: 10.1182/bloodadvances.2023009953
TP53 mutations identify high-risk events for peripheral T-cell lymphoma treated with CHOP-based chemotherapy
Abstract
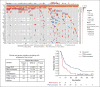
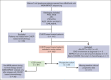
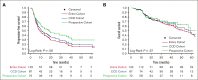
Nodal peripheral T-cell lymphomas (PTCL), the most common PTCLs, are generally treated with cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP)-based curative-intent chemotherapy. Recent molecular data have assisted in prognosticating these PTCLs, but most reports lack detailed baseline clinical characteristics and treatment courses. We retrospectively evaluated cases of PTCL treated with CHOP-based chemotherapy that had tumors sequenced by the Memorial Sloan Kettering Integrated Mutational Profiling of Actionable Cancer Targets next-generation sequencing panel to identify variables correlating with inferior survival. We identified 132 patients who met these criteria. Clinical factors correlating with an increased risk of progression (by multivariate analysis) included advanced-stage disease and bone marrow involvement. The only somatic genetic aberrancies correlating with inferior progression-free survival (PFS) were TP53 mutations and TP53/17p deletions. PFS remained inferior when stratifying by TP53 mutation status, with a median PFS of 4.5 months for PTCL with a TP53 mutation (n = 21) vs 10.5 months for PTCL without a TP53 mutation (n = 111). No TP53 aberrancy correlated with inferior overall survival (OS). Although rare (n = 9), CDKN2A-deleted PTCL correlated with inferior OS, with a median of 17.6 months vs 56.7 months for patients without CDKN2A deletions. This retrospective study suggests that patients with PTCL with TP53 mutations experience inferior PFS when treated with curative-intent chemotherapy, warranting prospective confirmation.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: W.T.J. received consulting fees from Myeloid Therapeutics. A.J.M. received research support from ADC Therapeutics, BeiGene, Miragen, Seattle Genetics, Merck, Bristol Myers Squibb, Incyte, and SecuraBio, and received honoraria from Affimed, Imbrium Therapeutics LP/Purdue, Janpix Ltd, Merck, Seattle Genetics, and Takeda. N.K. received research support from Seattle Genetics. A.D.Z. received consulting fees from Genentech/Roche, Gilead, Celgene, Janssen, Amgen, Novartis, Adaptive Biotechnology, MorphoSys, AbbVie, AstraZeneca, and MEI Pharma; received research funding from MEI Pharmaceuticals, Genentech/Roche, BeiGene, and NIH/NCI SPORE in Lymphoma (P50 CA192937-06A1); and served in data monitoring committees for BeiGene (Chair) and Bristol Myers Squibb/Celgene/Juno. M.L.P. received advisory/consulting fees from Novartis, Synthekine, BeiGene, Kite, and MustangBio. M.J.M. received honoraria from Genentech, Roche, GlaxoSmithKline, Bayer, Pharmacyclics, Janssen, Seattle Genetics, Immunovaccine Technologies, Takeda, and Epizyme; received advisory/consulting fees from Genentech, Bayer, Merck, Juno Therapeutics, Roche, Teva, Rocket Medical, Seattle Genetics, Daiichi Sankyo, Takeda, and Epizyme; and received research funding from Genentech, Roche, GlaxoSmithKline, IGM Biosciences, Bayer, Pharmacyclics, Janssen, Rocket Medical, Seattle Genetics, and Immunovaccine Technologies. A.N. received research funding from Pharmacyclics/AbbVie, Kite/Gilead, and Cornerstone; received consulting fees from Janssen, Morphosys, Cornerstone, Epizyme, EUSA, TG therapeutics, ADC Therapeutics, and AstraZeneca; and received honoraria from Pharmacyclics/AbbVie. P.C.C. holds stock/stock options in Bristol Myers Squibb, Johnson & Johnson, Pfizer, AstraZeneca, GlaxoSmithKline, and Novartis. C.L.B. is employed by Genentech. A.K. received research funding from AbbVie Pharmaceuticals, Adaptive Biotechnologies, Celgene, Pharmacyclics, Seattle Genetics, AstraZeneca, and Loxo Oncology/Lilly, and served in an advisory role for Celgene, Genentech, Kite Pharmaceuticals, Loxo Oncology/Lilly, and AstraZeneca. C.S.S. provided consulting for Juno Therapeutics, Sanofi-Genzyme, Spectrum Pharmaceuticals, Novartis, Genmab, Precision Biosciences, Kite/Gilead, Celgene/Bristol Myers Squibb, Gamida Cell, Karyopharm Therapeutics, Ono Pharmaceuticals, MorphoSys, CSL Behring, Syncopation Life Sciences, CRISPR Therapeutics, and GlaxoSmithKline, and received research funding from Juno Therapeutics, Celgene/Bristol Myers Squibb, Precision Biosciences, Actinium Pharmaceuticals, and Sanofi-Genzyme. L.F. received research funding and consulting fees from Genmab, AbbVie, and Roche/Genentech; received honoraria from and served on advisory boards for ADC Therapeutics, Seattle Genetics, and AstraZeneca. J.K.L. received consulting fees from TG Therapeutics and Epizyme. S.A.V. served on advisory board for Immunai, and received consulting fees from ADC Therapeutics and Koch Disruptive Technologies. G.S. served on advisory boards for and received consulting fees from AbbVie, Bayer, BeiGene, Bristol Myers Squibb/Celgene, Epizyme, Genentech/Roche, Genmab, Incyte, Ipsen, Janssen, Kite/Gilead, Loxo, Miltenyi, Molecular Partners, MorphoSys, Nordic Nanovector, Novartis, Rapt, Regeneron, and Takeda, and owns shares in Owkin. A.D. provided consulting for Incyte, EUSA Pharma, and Loxo, and received research support from Roche and Takeda. S.M.H. received research funding from ADC Therapeutics, Affimed, Aileron, Celgene, CRISPR Therapeutics, Daiichi Sankyo, Forty Seven Inc, Kyowa Hakko Kirin, Millennium/Takeda, Seattle Genetics, Trillium Therapeutics, and Verastem/Secura Bio, and received consulting fees from Acrotech Biopharma, ADC Therapeutics, Astex, Auxilus Pharma, Merck, C4 Therapeutics, Celgene, Cimieo Therapeutics, Daiichi Sankyo, Janssen, Kura Oncology, Kyowa Hakko Kirin, Myeloid Therapeutics, ONO Pharmaceuticals, Seattle Genetics, Secura Bio, Shoreline Biosciences Inc, Takeda, Trillium Therapeutics, Tubulis, Verastem/Secura Bio, Vividion Therapeutics, and Yingli Pharma Ltd. The remaining authors declare no competing financial interests.
A.M.H. is a retired member of the faculty of Memorial Sloan Kettering Cancer Center, New York, NY.
The current affiliation for C.L.B. is Genentech, South San Francisco, CA.
Figures



References
-
- Vose J, Armitage J, Weisenburger D, International T-Cell Lymphoma Project International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;26(25):4124–4130. - PubMed
-
- Ellin F, Landstrom J, Jerkeman M, Relander T. Real-world data on prognostic factors and treatment in peripheral T-cell lymphomas: a study from the Swedish Lymphoma Registry. Blood. 2014;124(10):1570–1577. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

