Epidemiological and Genetic Characteristics of Clinical Carbapenem-Resistant Pseudomonas aeruginosa Strains in Guangdong Province, China
- PMID: 37078855
- PMCID: PMC10269565
- DOI: 10.1128/spectrum.04261-22
Epidemiological and Genetic Characteristics of Clinical Carbapenem-Resistant Pseudomonas aeruginosa Strains in Guangdong Province, China
Abstract
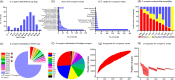
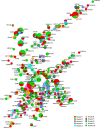
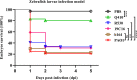
Carbapenem-resistant Pseudomonas aeruginosa (CRPA) is a bacterial pathogen that may cause serious drug-resistant infections that are potentially fatal. To investigate the genetic characteristics of these organisms, we tested 416 P. aeruginosa strains recovered from 12 types of clinical samples collected in 29 different hospital wards in 10 hospitals in Guangdong Province, China, from 2017 to 2020. These strains were found to belong to 149 known sequence types (STs) and 72 novel STs, indicating that transmission of these strains involved multiple routes. A high rate of resistance to imipenem (89.4%) and meropenem (79.4%) and a high prevalence of pathogenic serotypes (76.4%) were observed among these strains. Six STs of global high-risk clones (HiRiCs) and a novel HiRiC strains, ST1971, which exhibited extensive drug resistance, were identified. Importantly, ST1971 HiRiC, which was unique in China, also exhibited high virulence, which alarmed the further surveillance on this highly virulent and highly resistant clone. Inactivation of the oprD gene and overexpression of efflux systems were found to be mainly responsible for carbapenem resistance in these strains; carriage of metallo-β-lactamase (MBL)-encoding genes was less common. Interestingly, frameshift mutations (49.0%) and introduction of a stop codon (22.4%) into the oprD genes were the major mechanisms of imipenem resistance. On the other hand, expression of the MexAB-OprM efflux pump and MBL-encoding genes were mechanisms of resistance in >70% of meropenem-resistant strains. The findings presented here provide insights into the development of effective strategies for control of worldwide dissemination of CRPA. IMPORTANCE Carbapenem-resistant Pseudomonas aeruginosa (CRPA) is a major concern in clinical settings worldwide, yet few genetic and epidemiological studies on CRPA strains have been performed in China. Here, we sequence and analyze the genomes of 416 P. aeruginosa strains from hospitals in China to elucidate the genetic, phenotypic, and transmission characteristics of CRPA strains and to identify the molecular signatures responsible for the observed increase in the prevalence of CRPA infections in China. These findings may provide new insight into the development of effective strategies for worldwide control of CRPA and minimize the occurrence of untreatable infections in clinical settings.
Keywords: Pseudomonas aeruginosa; carbapenem resistance; efflux-pump-encoding genes; molecular epidemiology; oprD.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Streeter K, Katouli M. 2016. Pseudomonas aeruginosa: a review of their pathogenesis and prevalence in clinical settings and the environment. https://www.researchgate.net/publication/317695013_Pseudomonas_aeruginos....
-
- Centers for Disease Control and Prevention. 2018. Pseudomonas aeruginosa in healthcare settings. CDC, Atlanta, GA. https://www.cdc.gov/hai/organisms/pseudomonas.html. Accessed 27 August 2018.
-
- Armijo LM, Wawrzyniec SJ, Kopciuch M, Brandt YI, Rivera AC, Withers NJ, Cook NC, Huber DL, Monson TC, Smyth HDC, Osiński M. 2020. Antibacterial activity of iron oxide, iron nitride, and tobramycin conjugated nanoparticles against Pseudomonas aeruginosa biofilms. J Nanobiotechnol 18:1–27. doi:10.1186/s12951-020-0588-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

