MARCO Inhibits Porcine Reproductive and Respiratory Syndrome Virus Infection through Intensifying Viral GP5-Induced Apoptosis
- PMID: 37078873
- PMCID: PMC10269733
- DOI: 10.1128/spectrum.04753-22
MARCO Inhibits Porcine Reproductive and Respiratory Syndrome Virus Infection through Intensifying Viral GP5-Induced Apoptosis
Abstract
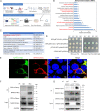
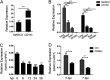
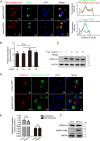
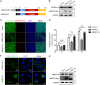
Studying viral glycoprotein-host membrane protein interactions contributes to the discovery of novel cell receptors or entry facilitators for viruses. Glycoprotein 5 (GP5), which is a major envelope protein of porcine reproductive and respiratory syndrome virus (PRRSV) virions, is a key target for the control of the virus. Here, the macrophage receptor with collagenous structure (MARCO), which is a member of the scavenger receptor family, was identified as one of the host interactors of GP5 through a DUALmembrane yeast two-hybrid screening. MARCO was specifically expressed on porcine alveolar macrophages (PAMs), and PRRSV infection downregulated MARCO expression both in vitro and in vivo. MARCO was not involved in viral adsorption and internalization processes, indicating that MARCO may not be a PRRSV-entry facilitator. Contrarily, MARCO served as a host restriction factor for PRRSV. The knockdown of MARCO in PAMs enhanced PRRSV proliferation, whereas overexpression suppressed viral proliferation. The N-terminal cytoplasmic region of MARCO was responsible for its inhibitory effect on PRRSV. Further, we found that MARCO was a proapoptotic factor in PRRSV-infected PAMs. MARCO knockdown weakened virus-induced apoptosis, whereas overexpression aggravated apoptosis. MARCO aggravated GP5-induced apoptosis, which may result in its proapoptotic function in PAMs. The interaction between MARCO and GP5 may contribute to the intensified apoptosis induced by GP5. Additionally, the inhibition of apoptosis during PRRSV infection weakened the antiviral function of MARCO, suggesting that MARCO inhibits PRRSV through the regulation of apoptosis. Taken together, the results of this study reveal a novel antiviral mechanism of MARCO and suggest a molecular basis for the potential development of therapeutics against PRRSV. IMPORTANCE Porcine reproductive and respiratory syndrome virus (PRRSV) has been one of the most serious threats to the global swine industry. Glycoprotein 5 (GP5) exposed on the surface of PRRSV virions is a major glycoprotein, and it is involved in viral entry into host cells. A macrophage receptor with collagenous structure (MARCO), which is a member of the scavenger receptor family, was identified to interact with PRRSV GP5 in a DUALmembrane yeast two-hybrid screening. Further investigation demonstrated that MARCO may not serve as a potential receptor to mediate PRRSV entry. Instead, MARCO was a host restriction factor for the virus, and the N-terminal cytoplasmic region of MARCO was responsible for its anti-PRRSV effect. Mechanistically, MARCO inhibited PRRSV infection through intensifying virus-induced apoptosis in PAMs. The interaction between MARCO and GP5 may contribute to GP5-induced apoptosis. Our work reveals a novel antiviral mechanism of MARCO and advances the development of control strategies for the virus.
Keywords: MARCO; apoptosis; glycoprotein 5; porcine reproductive and respiratory syndrome virus; yeast two-hybrid screening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

