PAReTT: A Python Package for the Automated Retrieval and Management of Divergence Time Data from the TimeTree Resource for Downstream Analyses
- PMID: 37079046
- PMCID: PMC10277261
- DOI: 10.1007/s00239-023-10106-3
PAReTT: A Python Package for the Automated Retrieval and Management of Divergence Time Data from the TimeTree Resource for Downstream Analyses
Abstract
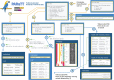
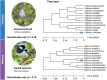
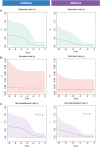
Evolutionary processes happen gradually over time and are, thus, considered time dependent. In addition, several evolutionary processes are either adaptations to local habitats or changing habitats, otherwise restricted thereby. Since evolutionary processes driving speciation take place within the landscape of environmental and temporal bounds, several published studies have aimed at providing accurate, fossil-calibrated, estimates of the divergence times of both extant and extinct species. Correct calibration is critical towards attributing evolutionary adaptations and speciation both to the time and paleogeography that contributed to it. Data from more than 4000 studies and nearly 1,50,000 species are available from a central TimeTree resource and provide opportunities of retrieving divergence times, evolutionary timelines, and time trees in various formats for most vertebrates. These data greatly enhance the ability of researchers to investigate evolution. However, there is limited functionality when studying lists of species that require batch retrieval. To overcome this, a PYTHON package termed Python-Automated Retrieval of TimeTree data (PAReTT) was created to facilitate a biologist-friendly interaction with the TimeTree resource. Here, we illustrate the use of the package through three examples that includes the use of timeline data, time tree data, and divergence time data. Furthermore, PAReTT was previously used in a meta-analysis of candidate genes to illustrate the relationship between divergence times and candidate genes of migration. The PAReTT package is available for download from GitHub or as a pre-compiled Windows executable, with extensive documentation on the package available on GitHub wiki pages regarding dependencies, installation, and implementation of the various functions.
Keywords: Divergence time; Diversification rate; PAReTT; PYTHON; Time trees; Timelines.
© 2023. The Author(s).
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures



Similar articles
-
TimeTree: A Resource for Timelines, Timetrees, and Divergence Times.Mol Biol Evol. 2017 Jul 1;34(7):1812-1819. doi: 10.1093/molbev/msx116. Mol Biol Evol. 2017. PMID: 28387841
-
TimeTree 5: An Expanded Resource for Species Divergence Times.Mol Biol Evol. 2022 Aug 6;39(8):msac174. doi: 10.1093/molbev/msac174. Online ahead of print. Mol Biol Evol. 2022. PMID: 35932227 Free PMC article.
-
Data-driven speciation tree prior for better species divergence times in calibration-poor molecular phylogenies.Bioinformatics. 2021 Jul 12;37(Suppl_1):i102-i110. doi: 10.1093/bioinformatics/btab307. Bioinformatics. 2021. PMID: 34252953 Free PMC article.
-
The fossil record and estimating divergence times between lineages: maximum divergence times and the importance of reliable phylogenies.J Mol Evol. 1990 May;30(5):400-8. doi: 10.1007/BF02101112. J Mol Evol. 1990. PMID: 2111853 Review.
-
Vicariance and dispersal in southern hemisphere freshwater fish clades: a palaeontological perspective.Biol Rev Camb Philos Soc. 2019 Apr;94(2):662-699. doi: 10.1111/brv.12473. Epub 2018 Oct 19. Biol Rev Camb Philos Soc. 2019. PMID: 30338909 Review.
Cited by
-
Phenotypic correlates between clock genes and phenology among populations of Diederik cuckoo, Chrysococcyx caprius.Ecol Evol. 2024 Jul 31;14(8):e70117. doi: 10.1002/ece3.70117. eCollection 2024 Aug. Ecol Evol. 2024. PMID: 39091329 Free PMC article.
References
-
- Barker K, Burns KJ, Klicka J, et al. New insights into new world biogeography: an integrated view from the phylogeny of blackbirds, cardinals, sparrows, tanagers, warblers, and allies. Auk. 2015;132:333–348. doi: 10.1642/AUK-14-110.1. - DOI
-
- BirdLife International, Handbook of the Birds of the World (2021) Bird species distribution maps of the world. Version 2021.1. http://datazone.birdlife.org/species/requestdis. Accessed 9 Dec 2021
-
- Carr J (2021) Mantel. PYTHON package version 2.1.0. https://github.com/jwcarr/mantel. Accessed 21 Apr 2022
-
- Castiglione S, Mondanaro A, Melchionna M, et al. Diversification rates and the evolution of species range size frequency distribution. Front Ecol Evol. 2017;5:147. doi: 10.3389/FEVO.2017.00147/BIBTEX. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

