High-throughput proteomic analysis reveals systemic dysregulation in virally suppressed people living with HIV
- PMID: 37079385
- PMCID: PMC10393229
- DOI: 10.1172/jci.insight.166166
High-throughput proteomic analysis reveals systemic dysregulation in virally suppressed people living with HIV
Abstract
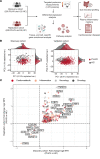
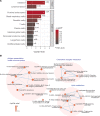
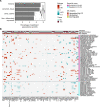
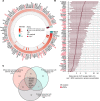
BACKGROUNDPeople living with HIV (PLHIV) receiving antiretroviral therapy (ART) exhibit persistent immune dysregulation and microbial dysbiosis, leading to development of cardiovascular diseases (CVDs). We initially compared plasma proteomic profiles between 205 PLHIV and 120 healthy control participants (HCs) and validated the results in an independent cohort of 639 PLHIV and 99 HCs. Differentially expressed proteins (DEPs) were then associated to microbiome data. Finally, we assessed which proteins were linked with CVD development in PLHIV.METHODSProximity extension assay technology was used to measure 1,472 plasma proteins. Markers of systemic inflammation (C-reactive protein, D-dimer, IL-6, soluble CD14, and soluble CD163) and microbial translocation (IFABP) were measured by ELISA, and gut bacterial species were identified using shotgun metagenomic sequencing. Baseline CVD data were available for all PLHIV, and 205 PLHIV were recorded for development of CVD during a 5-year follow-up.RESULTSPLHIV receiving ART had systemic dysregulation of protein concentrations, compared with HCs. Most of the DEPs originated from the intestine and lymphoid tissues and were enriched in immune- and lipid metabolism-related pathways. DEPs originating from the intestine were associated with specific gut bacterial species. Finally, we identified upregulated proteins in PLHIV (GDF15, PLAUR, RELT, NEFL, COL6A3, and EDA2R), unlike most markers of systemic inflammation, associated with the presence and risk of developing CVD during 5-year follow-up.CONCLUSIONOur findings suggest a systemic dysregulation of protein concentrations in PLHIV; some proteins were associated with CVD development. Most DEPs originated from the gut and were related to specific gut bacterial species.TRIAL REGISTRATIONClinicalTrials.gov NCT03994835.FUNDINGAIDS-fonds (P-29001), ViiV healthcare grant (A18-1052), Spinoza Prize (NWO SPI94-212), European Research Council (ERC) Advanced grant (grant 833247), and Indonesia Endowment Fund for Education.
Keywords: AIDS/HIV; Bioinformatics; Cardiovascular disease; Innate immunity.
Conflict of interest statement
Figures
References
-
- Croxford S, et al. Mortality and causes of death in people diagnosed with HIV in the era of highly active antiretroviral therapy compared with the general population: an analysis of a national observational cohort. Lancet Public Health. 2017;2(1):e35–e46. doi: 10.1016/S2468-2667(16)30020-2. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

