Extensive genome analysis identifies novel plasmid families in Clostridium perfringens
- PMID: 37079454
- PMCID: PMC10210947
- DOI: 10.1099/mgen.0.000995
Extensive genome analysis identifies novel plasmid families in Clostridium perfringens
Abstract
Globally, the anaerobic bacterium Clostridium perfringens causes severe disease in a wide array of hosts; however, C. perfringens strains are also carried asymptomatically. Accessory genes are responsible for much of the observed phenotypic variation and virulence within this species, with toxins frequently encoded on conjugative plasmids and many isolates carrying up to 10 plasmids. Despite this unusual biology, current genomic analyses have largely excluded isolates from healthy hosts or environmental sources. Accessory genomes, including plasmids, also have often been excluded from broader scale phylogenetic investigations. Here we interrogate a comprehensive collection of 464 C. perfringens genomes and identify the first putative non-conjugative enterotoxin (CPE)-encoding plasmids and a putative novel conjugative locus (Bcp) with sequence similarity to a locus reported from Clostridium botulinum. We sequenced and archived 102 new C. perfringens genomes, including those from rarely sequenced toxinotype B, C, D and E isolates. Long-read sequencing of 11 C. perfringens strains representing all toxinotypes (A-G) identified 55 plasmids from nine distinct plasmid groups. Interrogation of the 464 genomes in this collection identified 1045 plasmid-like contigs from the nine plasmid families, with a wide distribution across the C. perfringens isolates. Plasmids and plasmid diversity play an essential role in C. perfringens pathogenicity and broader biology. We have expanded the C. perfringens genome collection to include temporal, spatial and phenotypically diverse isolates including those carried asymptomatically in the gastrointestinal microbiome. This analysis has resulted in the identification of novel C. perfringens plasmids whilst providing a comprehensive understanding of species diversity.
Keywords: Clostridium perfringens; genome; horizontal gene transfer; phylogeny; plasmids; toxins.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

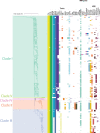
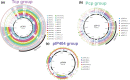



Similar articles
-
NetF-producing Clostridium perfringens: Clonality and plasmid pathogenicity loci analysis.Infect Genet Evol. 2017 Apr;49:32-38. doi: 10.1016/j.meegid.2016.12.028. Epub 2017 Jan 3. Infect Genet Evol. 2017. PMID: 28062388
-
pCP13, a representative of a new family of conjugative toxin plasmids in Clostridium perfringens.Plasmid. 2019 Mar;102:37-45. doi: 10.1016/j.plasmid.2019.02.002. Epub 2019 Feb 18. Plasmid. 2019. PMID: 30790588
-
Plasmid Characterization and Chromosome Analysis of Two netF+ Clostridium perfringens Isolates Associated with Foal and Canine Necrotizing Enteritis.PLoS One. 2016 Feb 9;11(2):e0148344. doi: 10.1371/journal.pone.0148344. eCollection 2016. PLoS One. 2016. PMID: 26859667 Free PMC article.
-
Genomic analyses of Clostridium perfringens isolates from five toxinotypes.Res Microbiol. 2015 May;166(4):255-63. doi: 10.1016/j.resmic.2014.10.003. Epub 2014 Oct 16. Res Microbiol. 2015. PMID: 25445567 Review.
-
Toxin plasmids of Clostridium perfringens.Microbiol Mol Biol Rev. 2013 Jun;77(2):208-33. doi: 10.1128/MMBR.00062-12. Microbiol Mol Biol Rev. 2013. PMID: 23699255 Free PMC article. Review.
Cited by
-
The biology and pathogenicity of Clostridium perfringens type F: a common human enteropathogen with a new(ish) name.Microbiol Mol Biol Rev. 2024 Sep 26;88(3):e0014023. doi: 10.1128/mmbr.00140-23. Epub 2024 Jun 12. Microbiol Mol Biol Rev. 2024. PMID: 38864615 Free PMC article. Review.
-
Clostridium perfringens-Opportunistic Foodborne Pathogen, Its Diversity and Epidemiological Significance.Pathogens. 2023 May 26;12(6):768. doi: 10.3390/pathogens12060768. Pathogens. 2023. PMID: 37375458 Free PMC article. Review.
-
Preparation and Application of Clostridium perfringens Alpha Toxin Nanobodies.Vet Sci. 2024 Aug 19;11(8):381. doi: 10.3390/vetsci11080381. Vet Sci. 2024. PMID: 39195835 Free PMC article.
-
Comparative genomic analysis of food-animal-derived and human-derived Clostridium perfringens isolates from markets in Shandong, China.Front Microbiol. 2025 Apr 1;16:1543511. doi: 10.3389/fmicb.2025.1543511. eCollection 2025. Front Microbiol. 2025. PMID: 40236475 Free PMC article.
-
Unveiling the pathogenic mechanisms of Clostridium perfringens toxins and virulence factors.Emerg Microbes Infect. 2024 Dec;13(1):2341968. doi: 10.1080/22221751.2024.2341968. Epub 2024 Apr 27. Emerg Microbes Infect. 2024. PMID: 38590276 Free PMC article. Review.
References
-
- Sarker MR, Carman RJ, McClane BA. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol Microbiol. 1999;33:946–958. doi: 10.1046/j.1365-2958.1999.01534.x. - DOI - PubMed
-
- Awad MM, Bryant AE, Stevens DL, Rood JI. Virulence studies on chromosomal alpha-toxin and theta-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of alpha-toxin in Clostridium perfringens-mediated gas gangrene. Mol Microbiol. 1995;15:191–202. doi: 10.1111/j.1365-2958.1995.tb02234.x. - DOI - PubMed
-
- Blackwell TE, Butler DG, Prescott JF, Wilcock BP. Differences in signs and lesions in sheep and goats with enterotoxemia induced by intraduodenal infusion of Clostridium perfringens type D. Am J Vet Res. 1991;52:1147–1152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

