CONNECTOR, fitting and clustering of longitudinal data to reveal a new risk stratification system
- PMID: 37079732
- PMCID: PMC10159654
- DOI: 10.1093/bioinformatics/btad201
CONNECTOR, fitting and clustering of longitudinal data to reveal a new risk stratification system
Abstract
Motivation: The transition from evaluating a single time point to examining the entire dynamic evolution of a system is possible only in the presence of the proper framework. The strong variability of dynamic evolution makes the definition of an explanatory procedure for data fitting and clustering challenging.
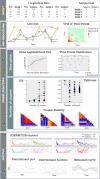
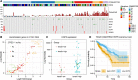
Results: We developed CONNECTOR, a data-driven framework able to analyze and inspect longitudinal data in a straightforward and revealing way. When used to analyze tumor growth kinetics over time in 1599 patient-derived xenograft growth curves from ovarian and colorectal cancers, CONNECTOR allowed the aggregation of time-series data through an unsupervised approach in informative clusters. We give a new perspective of mechanism interpretation, specifically, we define novel model aggregations and we identify unanticipated molecular associations with response to clinically approved therapies.
Availability and implementation: CONNECTOR is freely available under GNU GPL license at https://qbioturin.github.io/connector and https://doi.org/10.17504/protocols.io.8epv56e74g1b/v1.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
L.T. has received research grants from Menarini, Merck KGaA, Merus, Pfizer, Servier and Symphogen. The other authors declare no conflicts.
Figures

References
-
- Bertotti A, Migliardi G, Galimi F. et al. A molecularly annotated platform of patient-derived xenografts (”xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov 2011;1:508–23. 10.1158/2159-8290.CD-11-0109. - DOI - PubMed
-
- Davies DL, Bouldin DW.. A cluster separation measure. IEEE Trans Pattern Anal Mach Intell 1979;1:224–7. - PubMed

