SituSeq: an offline protocol for rapid and remote Nanopore 16S rRNA amplicon sequence analysis
- PMID: 37081077
- PMCID: PMC10119094
- DOI: 10.1038/s43705-023-00239-3
SituSeq: an offline protocol for rapid and remote Nanopore 16S rRNA amplicon sequence analysis
Abstract
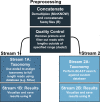
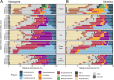
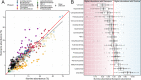
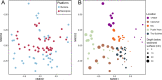
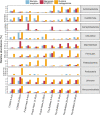
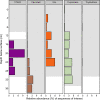
Microbiome analysis through 16S rRNA gene sequencing is a crucial tool for understanding the microbial ecology of any habitat or ecosystem. However, workflows require large equipment, stable internet, and extensive computing power such that most of the work is performed far away from sample collection in both space and time. Performing amplicon sequencing and analysis at sample collection would have positive implications in many instances including remote fieldwork and point-of-care medical diagnoses. Here we present SituSeq, an offline and portable workflow for the sequencing and analysis of 16S rRNA gene amplicons using Nanopore sequencing and a standard laptop computer. SituSeq was validated by comparing Nanopore 16S rRNA gene amplicons, Illumina 16S rRNA gene amplicons, and Illumina metagenomes, sequenced using the same environmental DNA. Comparisons revealed consistent community composition, ecological trends, and sequence identity across platforms. Correlation between the abundance of taxa in each taxonomic level in Illumina and Nanopore data sets was high (Pearson's r > 0.9), and over 70% of Illumina 16S rRNA gene sequences matched a Nanopore sequence with greater than 97% sequence identity. On board a research vessel on the open ocean, SituSeq was used to analyze amplicon sequences from deep sea sediments less than 2 h after sequencing, and 8 h after sample collection. The rapidly available results informed decisions about subsequent sampling in near real-time while the offshore expedition was still underway. SituSeq is a portable and user-friendly workflow that helps to bring the power of microbial genomics and diagnostics to many more researchers and situations.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
High accuracy meets high throughput for near full-length 16S ribosomal RNA amplicon sequencing on the Nanopore platform.PNAS Nexus. 2024 Oct 9;3(10):pgae411. doi: 10.1093/pnasnexus/pgae411. eCollection 2024 Oct. PNAS Nexus. 2024. PMID: 39386005 Free PMC article.
-
VITCOMIC2: visualization tool for the phylogenetic composition of microbial communities based on 16S rRNA gene amplicons and metagenomic shotgun sequencing.BMC Syst Biol. 2018 Mar 19;12(Suppl 2):30. doi: 10.1186/s12918-018-0545-2. BMC Syst Biol. 2018. PMID: 29560821 Free PMC article.
-
The Clinical Utility of Two High-Throughput 16S rRNA Gene Sequencing Workflows for Taxonomic Assignment of Unidentifiable Bacterial Pathogens in Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry.J Clin Microbiol. 2022 Jan 19;60(1):e0176921. doi: 10.1128/JCM.01769-21. Epub 2021 Nov 17. J Clin Microbiol. 2022. PMID: 34788113 Free PMC article.
-
Computational methods for 16S metabarcoding studies using Nanopore sequencing data.Comput Struct Biotechnol J. 2020 Jan 31;18:296-305. doi: 10.1016/j.csbj.2020.01.005. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32071706 Free PMC article. Review.
-
Beyond Basic Diversity Estimates-Analytical Tools for Mechanistic Interpretations of Amplicon Sequencing Data.Microorganisms. 2022 Oct 1;10(10):1961. doi: 10.3390/microorganisms10101961. Microorganisms. 2022. PMID: 36296237 Free PMC article. Review.
Cited by
-
Advancing animal tuberculosis surveillance using culture-independent long-read whole-genome sequencing.Front Microbiol. 2023 Nov 21;14:1307440. doi: 10.3389/fmicb.2023.1307440. eCollection 2023. Front Microbiol. 2023. PMID: 38075895 Free PMC article.
-
rCRUX: A Rapid and Versatile Tool for Generating Metabarcoding Reference libraries in R.Environ DNA. 2024 Jan;6(1):e489. doi: 10.1002/edn3.489. Epub 2023 Nov 29. Environ DNA. 2024. PMID: 38370872 Free PMC article.
-
Improved resolution of microbial diversity in deep-sea surface sediments using PacBio long-read 16S rRNA gene sequencing.mSphere. 2024 Dec 19;9(12):e0077024. doi: 10.1128/msphere.00770-24. Epub 2024 Nov 12. mSphere. 2024. PMID: 39530673 Free PMC article.
-
rCRUX: A Rapid and Versatile Tool for Generating Metabarcoding Reference libraries in R.bioRxiv [Preprint]. 2023 Jun 3:2023.05.31.543005. doi: 10.1101/2023.05.31.543005. bioRxiv. 2023. Update in: Environ DNA. 2024 Jan;6(1):e489. doi: 10.1002/edn3.489. PMID: 37397980 Free PMC article. Updated. Preprint.
-
NanoASV: a snakemake workflow for reproducible field-based Nanopore full-length 16S metabarcoding amplicon data analysis.Bioinformatics. 2025 Mar 4;41(3):btaf089. doi: 10.1093/bioinformatics/btaf089. Bioinformatics. 2025. PMID: 40111866 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

