Nuclear and cytoplasmic spatial protein quality control is coordinated by nuclear-vacuolar junctions and perinuclear ESCRT
- PMID: 37081164
- PMCID: PMC12349969
- DOI: 10.1038/s41556-023-01128-6
Nuclear and cytoplasmic spatial protein quality control is coordinated by nuclear-vacuolar junctions and perinuclear ESCRT
Abstract
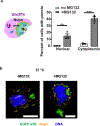
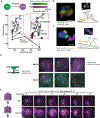
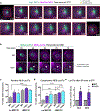
Effective protein quality control (PQC), essential for cellular health, relies on spatial sequestration of misfolded proteins into defined inclusions. Here we reveal the coordination of nuclear and cytoplasmic spatial PQC. Cytoplasmic misfolded proteins concentrate in a cytoplasmic juxtanuclear quality control compartment, while nuclear misfolded proteins sequester into an intranuclear quality control compartment (INQ). Particle tracking reveals that INQ and the juxtanuclear quality control compartment converge to face each other across the nuclear envelope at a site proximal to the nuclear-vacuolar junction marked by perinuclear ESCRT-II/III protein Chm7. Strikingly, convergence at nuclear-vacuolar junction contacts facilitates VPS4-dependent vacuolar clearance of misfolded cytoplasmic and nuclear proteins, the latter entailing extrusion of nuclear INQ into the vacuole. Finding that nuclear-vacuolar contact sites are cellular hubs of spatial PQC to facilitate vacuolar clearance of nuclear and cytoplasmic inclusions highlights the role of cellular architecture in proteostasis maintenance.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
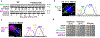

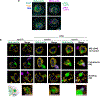
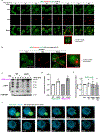

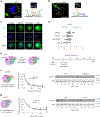
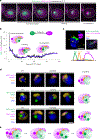

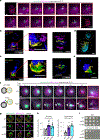


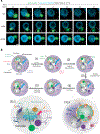
Comment in
-
A shared fate for nuclear and cytosolic inclusions.Nat Cell Biol. 2023 May;25(5):629-630. doi: 10.1038/s41556-023-01121-z. Nat Cell Biol. 2023. PMID: 37081163 No abstract available.
References
-
- Hartl FU, Bracher A & Hayer-Hartl M Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011). - PubMed
-
- Balchin D, Hayer-Hartl M & Hartl FU In vivo aspects of protein folding and quality control. Science 353, aac4354 (2016). - PubMed
-
- Sontag EM, Samant RS & Frydman J Mechanisms and functions of spatial protein quality control. Annu. Rev. Biochem. 20, 97–122 (2017). - PubMed
-
- Pohl C & Dikic I Cellular quality control by the ubiquitin–proteasome system and autophagy. Science 366, 818–822 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

