A positive feedback loop controls Toxoplasma chronic differentiation
- PMID: 37081202
- PMCID: PMC10520893
- DOI: 10.1038/s41564-023-01358-2
A positive feedback loop controls Toxoplasma chronic differentiation
Erratum in
-
Author Correction: A positive feedback loop controls Toxoplasma chronic differentiation.Nat Microbiol. 2024 Apr;9(4):1145. doi: 10.1038/s41564-023-01467-y. Nat Microbiol. 2024. PMID: 37582867 No abstract available.
Abstract
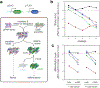
Successful infection strategies must balance pathogen amplification and persistence. In the obligate intracellular parasite Toxoplasma gondii this is accomplished through differentiation into dedicated cyst-forming chronic stages that avoid clearance by the host immune system. The transcription factor BFD1 is both necessary and sufficient for stage conversion; however, its regulation is not understood. In this study we examine five factors that are transcriptionally activated by BFD1. One of these is a cytosolic RNA-binding protein of the CCCH-type zinc-finger family, which we name bradyzoite formation deficient 2 (BFD2). Parasites lacking BFD2 fail to induce BFD1 and are consequently unable to fully differentiate in culture or in mice. BFD2 interacts with the BFD1 transcript under stress, and deletion of BFD2 reduces BFD1 protein levels but not messenger RNA abundance. The reciprocal effects on BFD2 transcription and BFD1 translation outline a positive feedback loop that enforces the chronic-stage gene-expression programme. Thus, our findings help explain how parasites both initiate and commit to chronic differentiation. This work provides new mechanistic insight into the regulation of T. gondii persistence, and can be exploited in the design of strategies to prevent and treat these key reservoirs of human infection.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
Figures
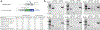


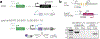

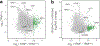






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

