Disentangling temporal associations in marine microbial networks
- PMID: 37081491
- PMCID: PMC10120119
- DOI: 10.1186/s40168-023-01523-z
Disentangling temporal associations in marine microbial networks
Abstract
Background: Microbial interactions are fundamental for Earth's ecosystem functioning and biogeochemical cycling. Nevertheless, they are challenging to identify and remain barely known. Omics-based censuses are helpful in predicting microbial interactions through the statistical inference of single (static) association networks. Yet, microbial interactions are dynamic and we have limited knowledge of how they change over time. Here, we investigate the dynamics of microbial associations in a 10-year marine time series in the Mediterranean Sea using an approach inferring a time-resolved (temporal) network from a single static network.
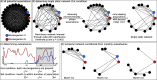
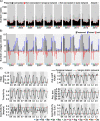
Results: A single static network including microbial eukaryotes and bacteria was built using metabarcoding data derived from 120 monthly samples. For the decade, we aimed to identify persistent, seasonal, and temporary microbial associations by determining a temporal network that captures the interactome of each individual sample. We found that the temporal network appears to follow an annual cycle, collapsing, and reassembling when transiting between colder and warmer waters. We observed higher association repeatability in colder than in warmer months. Only 16 associations could be validated using observations reported in literature, underlining our knowledge gap in marine microbial ecological interactions.
Conclusions: Our results indicate that marine microbial associations follow recurrent temporal dynamics in temperate zones, which need to be accounted for to better understand the functioning of the ocean microbiome. The constructed marine temporal network may serve as a resource for testing season-specific microbial interaction hypotheses. The applied approach can be transferred to microbiome studies in other ecosystems. Video Abstract.
Keywords: Association network; Microbial interactions; Microorganisms; Ocean; Plankton; Temporal network; Time series.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320:1034–1039. - PubMed
-
- DeLong EF. The microbial ocean from genomes to biomes. Nature. 2009;459:200–206. - PubMed
-
- Krabberød AK, Bjorbækmo MFM, Shalchian-Tabrizi K, Logares R. Exploring the oceanic microeukaryotic interactome with metaomics approaches. Aquat Microb Ecol. 2017;79:1–12.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

