Use of High-Resolution Geospatial and Genomic Data to Characterize Recent Tuberculosis Transmission, Botswana
- PMID: 37081530
- PMCID: PMC10124643
- DOI: 10.3201/eid2905.220796
Use of High-Resolution Geospatial and Genomic Data to Characterize Recent Tuberculosis Transmission, Botswana
Abstract
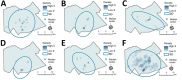
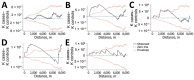
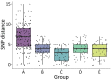
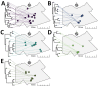
Combining genomic and geospatial data can be useful for understanding Mycobacterium tuberculosis transmission in high-burden tuberculosis (TB) settings. We performed whole-genome sequencing on M. tuberculosis DNA extracted from sputum cultures from a population-based TB study conducted in Gaborone, Botswana, during 2012-2016. We determined spatial distribution of cases on the basis of shared genotypes among isolates. We considered clusters of isolates with ≤5 single-nucleotide polymorphisms identified by whole-genome sequencing to indicate recent transmission and clusters of ≥10 persons to be outbreaks. We obtained both molecular and geospatial data for 946/1,449 (65%) participants with culture-confirmed TB; 62 persons belonged to 5 outbreaks of 10-19 persons each. We detected geospatial clustering in just 2 of those 5 outbreaks, suggesting heterogeneous spatial patterns. Our findings indicate that targeted interventions applied in smaller geographic areas of high-burden TB identified using integrated genomic and geospatial data might help interrupt TB transmission during outbreaks.
Keywords: Botswana; bacteria; geographic heterogeneity; infectious disease control; outbreaks; respiratory infections; spatial analysis; tuberculosis and other mycobacteria; whole-genome sequencing.
Figures




References
-
- World Health Organization. Global tuberculosis report 2021. [cited 2021 Oct 19]. https://www.who.int/publications-detail-redirect/9789240037021

