Not all exons are protein coding: Addressing a common misconception
- PMID: 37082142
- PMCID: PMC10112331
- DOI: 10.1016/j.xgen.2023.100296
Not all exons are protein coding: Addressing a common misconception
Abstract
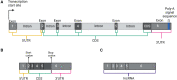
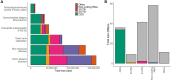
Exons are regions of DNA that are transcribed to RNA and retained after introns are spliced out. However, the term "exon" is often misused as synonymous to "protein coding," including in some literature and textbook definitions. In contrast, only a fraction of exonic sequences are protein coding (<30% in humans). Both exons and introns are also present in untranslated regions (UTRs) and non-coding RNAs. Misuse of the term exon is problematic, for example, "whole-exome sequencing" technology targets <25% of the human exome, primarily regions that are protein coding. Here, we argue for the importance of the original definition of an exon for making functional distinctions in genetics and genomics. Further, we recommend the use of clearer language referring to coding exonic regions and non-coding exonic regions. We propose the use of coding exome sequencing, or CES, to more appropriately describe sequencing approaches that target primarily protein-coding regions rather than all transcribed regions.
Keywords: UTRs; exome sequencing; exons; introns; non-coding RNA; splicing; untranslated regions.
© 2023 1.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Krebs J.E., Goldstein E.S., Kilpatrick S.T. Jones & Bartlett Publishers; 2009. Lewin’s GENES X.
-
- Gilbert W. Why genes in pieces? Nature. 1978;271:501. - PubMed
-
- Black D.L. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. - PubMed
-
- Adams J.M., Cory S. Untranslated nucleotide sequence at the 5’-end of R17 bacteriophage RNA. Nature. 1970;227:570–574. - PubMed
-
- Proudfoot N.J., Brownlee G.G. Sequence at the 3’ end of globin mRNA shows homology with immunoglobulin light chain mRNA. Nature. 1974;252:359–362. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

