Identification of key microRNAs regulating ELOVL6 and glioblastoma tumorigenesis
- PMID: 37082255
- PMCID: PMC10074970
- DOI: 10.1016/j.bbadva.2023.100078
Identification of key microRNAs regulating ELOVL6 and glioblastoma tumorigenesis
Abstract
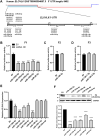
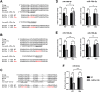
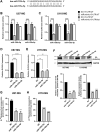
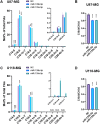
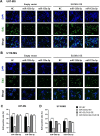
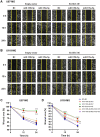
ELOVL fatty acid elongase 6 (ELOVL6) controls cellular fatty acid (FA) composition by catalyzing the elongation of palmitate (C16:0) to stearate (C18:0) and palmitoleate (C16:1n-7) to vaccinate (C18:1n-7). Although the transcriptional regulation of ELOVL6 has been well studied, the post-transcriptional regulation of ELOVL6 is not fully understood. Therefore, this study aims to evaluate the role of microRNAs (miRNAs) in regulating human ELOVL6. Bioinformatic analysis identified five putative miRNAs: miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, and miR-22-3p, which potentially bind ELOVL6 3'-untranslated region (UTR). Results from dual-luciferase assays revealed that these miRNAs downregulate ELOVL6 by directly interacting with the 3'-UTR of ELOVL6 mRNA. Moreover, miR-135b-5p and miR-135a-5p suppress cell proliferation and migration in glioblastoma multiforme cells by inhibiting ELOVL6 at the mRNA and protein levels. Taken together, our results provide novel regulatory mechanisms for ELOVL6 at the post-transcriptional level and identify potential candidates for the treatment of patients with glioblastoma multiforme.
Keywords: Cell proliferation; Fatty acid; Glioblastoma; MicroRNA; Migration.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that there is no conflict of interest associated with this manuscript.
Figures
References
-
- Guillou H., Zadravec D., Martin P.G., Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: insights from transgenic mice. Prog. Lipid Res. 2010;49:186–199. - PubMed
-
- Jakobsson A., Westerberg R., Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog. Lipid Res. 2006;45:237–249. - PubMed
-
- Moon Y.A., Shah N.A., Mohapatra S., Warrington J.A., Horton J.D. Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 2001;276:45358–45366. - PubMed
-
- Matsuzaka T., Shimano H., Yahagi N., Yoshikawa T., Amemiya-Kudo M., Hasty A.H., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Osuga J., Takahashi A., Yato S., Sone H., Ishibashi S., Yamada N. Cloning and characterization of a mammalian fatty acyl-CoA elongase as a lipogenic enzyme regulated by SREBPs. J. Lipid Res. 2002;43:911–920. - PubMed
LinkOut - more resources
Full Text Sources

