Selective SARS-CoV2 BA.2 escape of antibody Fc/Fc-receptor interactions
- PMID: 37082529
- PMCID: PMC10079316
- DOI: 10.1016/j.isci.2023.106582
Selective SARS-CoV2 BA.2 escape of antibody Fc/Fc-receptor interactions
Abstract
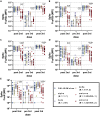
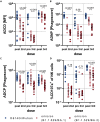
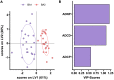
The number of mutations in the omicron (B.1.1.529) BA.1 variant of concern led to an unprecedented evasion of vaccine induced immunity. However, despite rise in global infections, severe disease did not increase proportionally and is likely linked to persistent recognition of BA.1 by T cells and non-neutralizing opsonophagocytic antibodies. Yet, the emergence of new sublineage BA.2, which is more transmissible than BA.1 despite relatively preserved neutralizing antibody responses, has raised the possibility that BA.2 may evade other vaccine-induced responses. Here, we comprehensively profiled the BNT162b2 vaccine-induced response to several VOCs, including omicron BA.1 and BA.2. While vaccine-induced immune responses were compromised against both omicron sublineages, vaccine-induced antibody isotype titers, and non-neutralizing Fc effector functions were attenuated to the omicron BA.2 spike compared to BA.1. Conversely, FcγR2a and FcγR2b binding was elevated to BA.2, albeit lower than BA.1 responses, potentially contributing to persistent protection against severity of disease.
Keywords: Immunology; Virology.
© 2023 The Author(s).
Conflict of interest statement
G.A. is a founder of Seromyx Systems and an equity holder in Leyden Labs. G.A. is on a leave of absence and is currently employed by Moderna Inc. D.H.B. is a co-inventor on provisional vaccine patents licensed to Janssen (63/121,482; 63/133,969; 63/135,182) and serves as a consultant to Pfizer. Y.C.B. is a consultant for Gorman Consulting. All other authors have nothing to declare.
Figures


Similar articles
-
The Biological Functions and Clinical Significance of SARS-CoV-2 Variants of Corcern.Front Med (Lausanne). 2022 May 20;9:849217. doi: 10.3389/fmed.2022.849217. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35669924 Free PMC article. Review.
-
Preserved recognition of Omicron spike following COVID-19 messenger RNA vaccination in pregnancy.Am J Obstet Gynecol. 2022 Sep;227(3):493.e1-493.e7. doi: 10.1016/j.ajog.2022.04.009. Epub 2022 Apr 14. Am J Obstet Gynecol. 2022. PMID: 35430229 Free PMC article.
-
Progressive loss of conserved spike protein neutralizing antibody sites in Omicron sublineages is balanced by preserved T cell immunity.Cell Rep. 2023 Aug 29;42(8):112888. doi: 10.1016/j.celrep.2023.112888. Epub 2023 Jul 31. Cell Rep. 2023. PMID: 37527039
-
Omicron variant Spike-specific antibody binding and Fc activity are preserved in recipients of mRNA or inactivated COVID-19 vaccines.Sci Transl Med. 2022 Apr 27;14(642):eabn9243. doi: 10.1126/scitranslmed.abn9243. Epub 2022 Apr 27. Sci Transl Med. 2022. PMID: 35289637 Free PMC article.
-
The humoral and cellular immune evasion of SARS-CoV-2 Omicron and sub-lineages.Virol Sin. 2022 Dec;37(6):786-795. doi: 10.1016/j.virs.2022.11.007. Epub 2022 Nov 23. Virol Sin. 2022. PMID: 36427646 Free PMC article. Review.
Cited by
-
Unpredicted Protective Function of Fc-Mediated Inhibitory Antibodies for HIV and SARS-CoV-2 Vaccines.J Infect Dis. 2025 Feb 4;231(1):e1-e9. doi: 10.1093/infdis/jiae464. J Infect Dis. 2025. PMID: 39302695 Free PMC article. Review.
-
Antibody Fc-binding profiles and ACE2 affinity to SARS-CoV-2 RBD variants.Med Microbiol Immunol. 2023 Aug;212(4):291-305. doi: 10.1007/s00430-023-00773-w. Epub 2023 Jul 21. Med Microbiol Immunol. 2023. PMID: 37477828 Free PMC article.
-
Anti-TNF therapy impairs both short- and long-term IgG responses after repeated vaccination.Allergy. 2025 Feb;80(2):423-439. doi: 10.1111/all.16241. Epub 2024 Jul 25. Allergy. 2025. PMID: 39049686 Free PMC article.
-
Diversity of immunization strongly impacts SARS-CoV-2 antibody function surrogates.NPJ Vaccines. 2025 Jul 29;10(1):175. doi: 10.1038/s41541-025-01226-6. NPJ Vaccines. 2025. PMID: 40730842 Free PMC article.
-
SARS-CoV-2 BA.4/5 infection triggers more cross-reactive FcγRIIIa signaling and neutralization than BA.1, in the context of hybrid immunity.J Virol. 2024 Jul 23;98(7):e0067824. doi: 10.1128/jvi.00678-24. Epub 2024 Jul 2. J Virol. 2024. PMID: 38953380 Free PMC article.
References
-
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., COVID-19 Genomics UK COG-UK Consortium, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. - DOI - PMC - PubMed
-
- VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Jr., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28:490–495. doi: 10.1038/s41591-021-01678-y. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

