C11orf21, a novel RUNX1 target gene, is down-regulated by RUNX1-ETO
- PMID: 37082605
- PMCID: PMC10074976
- DOI: 10.1016/j.bbadva.2022.100047
C11orf21, a novel RUNX1 target gene, is down-regulated by RUNX1-ETO
Abstract
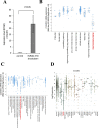
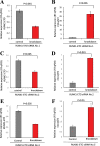
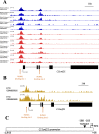
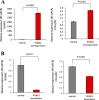
The fusion protein RUNX1-ETO is an oncogenic transcription factor generated by t(8;21) chromosome translocation, which is found in FAB-M2-type acute myeloid leukemia (AML). RUNX1-ETO is known to dysregulate the normal RUNX1 transcriptional network, which should involve essential factors for the onset of AML with t(8;21). In this study, we screened for possible transcriptional targets of RUNX1 by reanalysis of public data in silico, and identified C11orf21 as a novel RUNX1 target gene because its expression was down-regulated in the presence of RUNX1-ETO. The expression level of C11orf21 was low in AML patient samples with t(8;21) and in Kasumi-1 cells, which carry RUNX1-ETO. Knockdown of RUNX1-ETO in Kasumi-1 cells restored C11orf21 expression, whereas overexpression of RUNX1 up-regulated C11orf21 expression. In addition, knockdown of RUNX1 in other human leukemia cells without RUNX-ETO, such as K562, led to a decrease in C11orf21 expression. Of note, the C11orf21 promoter sequence contains a consensus sequence for RUNX1 binding and it was activated by exogenously expressed RUNX1 based on our luciferase reporter assay. This luciferase signal was trans-dominantly suppressed by RUNX1-ETO and site-directed mutagenesis of the consensus site abrogated the reporter activity. This study demonstrated that C11orf21 is a novel transcriptional target of RUNX1 and RUNX1-ETO suppressed C11orf21 transcription in t(8;21) AML. Thus, through this in silico approach, we identified a novel transcriptional target of RUNX1, and the depletion of C11orf21, the target gene, may be associated with the onset of t(8;21) AML.
Keywords: C11orf21; RNA-seq; RUNX1; RUNX1-ETO; t(8:21) AML.
© 2022 The Authors.
Conflict of interest statement
The authors declare no conflict of interest. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
The RUNX1-ETO fusion protein trans-activates c-KIT expression by recruiting histone acetyltransferase P300 on its promoter.FEBS J. 2019 Mar;286(5):901-912. doi: 10.1111/febs.14751. Epub 2019 Feb 15. FEBS J. 2019. PMID: 30637949
-
Stable depletion of RUNX1-ETO in Kasumi-1 cells induces expression and enhanced proteolytic activity of Cathepsin G and Neutrophil Elastase.PLoS One. 2019 Dec 11;14(12):e0225977. doi: 10.1371/journal.pone.0225977. eCollection 2019. PLoS One. 2019. PMID: 31826021 Free PMC article.
-
Repression of vascular endothelial growth factor expression by the runt-related transcription factor 1 in acute myeloid leukemia.Cancer Res. 2011 Apr 1;71(7):2761-71. doi: 10.1158/0008-5472.CAN-10-0402. Epub 2011 Mar 29. Cancer Res. 2011. PMID: 21447743
-
AML-1-ETO-Mediated erythroid inhibition: new paradigms for differentiation blockade by a leukemic fusion protein.Crit Rev Eukaryot Gene Expr. 2005;15(3):207-16. doi: 10.1615/critreveukargeneexpr.v15.i3.30. Crit Rev Eukaryot Gene Expr. 2005. PMID: 16390317 Review.
-
RUNX1 and RUNX1-ETO: roles in hematopoiesis and leukemogenesis.Front Biosci (Landmark Ed). 2012 Jan 1;17(3):1120-39. doi: 10.2741/3977. Front Biosci (Landmark Ed). 2012. PMID: 22201794 Free PMC article. Review.
Cited by
-
Application of CRISPR/Cas9 Technology in Cancer Treatment: A Future Direction.Curr Oncol. 2023 Feb 6;30(2):1954-1976. doi: 10.3390/curroncol30020152. Curr Oncol. 2023. PMID: 36826113 Free PMC article. Review.
-
BCL11B promotes T-cell acute lymphoblastic leukaemia cell survival via the XRCC5/C11ORF21 axis.Clin Transl Med. 2024 Feb;14(2):e1580. doi: 10.1002/ctm2.1580. Clin Transl Med. 2024. PMID: 38317587 Free PMC article. No abstract available.
-
The combination of venetoclax and quercetin exerts a cytotoxic effect on acute myeloid leukemia.Sci Rep. 2024 Nov 2;14(1):26418. doi: 10.1038/s41598-024-78221-9. Sci Rep. 2024. PMID: 39488609 Free PMC article.

