Association of frequent hypermethylation with high grade histological subtype in lung adenocarcinoma
- PMID: 37082886
- PMCID: PMC10323093
- DOI: 10.1111/cas.15817
Association of frequent hypermethylation with high grade histological subtype in lung adenocarcinoma
Abstract
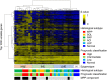
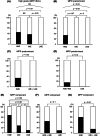
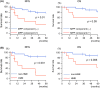
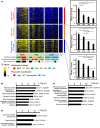
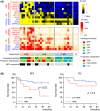
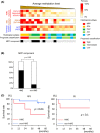
Lung adenocarcinoma is classified morphologically into five histological subtypes according to the WHO classification. While each histological subtype correlates with a distinct prognosis, the molecular basis has not been fully elucidated. Here we conducted DNA methylation analysis of 30 lung adenocarcinoma cases annotated with the predominant histological subtypes and three normal lung cases using the Infinium BeadChip. Unsupervised hierarchical clustering analysis revealed three subgroups with different methylation levels: high-, intermediate-, and low-methylation epigenotypes (HME, IME, and LME). Micropapillary pattern (MPP)-predominant cases and those with MPP components were significantly enriched in HME (p = 0.02 and p = 0.03, respectively). HME cases showed a significantly poor prognosis for recurrence-free survival (p < 0.001) and overall survival (p = 0.006). We identified 365 HME marker genes specifically hypermethylated in HME cases with enrichment of "cell morphogenesis" related genes; 305 IME marker genes hypermethylated in HME and IME, but not in LME, with enrichment "embryonic organ morphogenesis"-related genes; 257 Common marker genes hypermethylated commonly in all cancer cases, with enrichment of "regionalization"-related genes. We extracted surrogate markers for each epigenotype and designed pyrosequencing primers for five HME markers (TCERG1L, CXCL12, FAM181B, HOXA11, GAD2), three IME markers (TBX18, ZNF154, NWD2) and three Common markers (SCT, GJD2, BARHL2). DNA methylation profiling using Infinium data was validated by pyrosequencing, and HME cases defined by pyrosequencing results also showed the worse recurrence-free survival. In conclusion, lung adenocarcinomas are stratified into subtypes with distinct DNA methylation levels, and the high-methylation subtype correlated with MPP-predominant cases and those with MPP components and showed a poor prognosis.
Keywords: DNA methylation; high-grade subtype; histological subtype; lung adenocarcinoma; micropapillary component.
© 2023 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
A.K. is an Associate Editor of Cancer Science, and all other authors have no conflict of interest.
Figures
References
-
- Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBCAN sources and methods. Int J Cancer. 2019;144:1941‐1953. - PubMed
-
- Travis WD, Brambilla E, Burke A. WHO Classification of the Tumours of the Lung, Pleura, Thymus and Heart (eds 4). IARC Press; 2015. - PubMed
-
- Warth A, Muley T, Meister M, et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage‐independent predictor of survival. J Clin Oncol. 2012;3013:1438‐1446. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

