Non‑coding RNAs: Role of miRNAs and lncRNAs in the regulation of autophagy in hepatocellular carcinoma (Review)
- PMID: 37083063
- PMCID: PMC10170481
- DOI: 10.3892/or.2023.8550
Non‑coding RNAs: Role of miRNAs and lncRNAs in the regulation of autophagy in hepatocellular carcinoma (Review)
Abstract
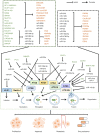
The term autophagy describes a process that supports nutrient cycling and metabolic adaptation that is accomplished via multistep lysosomal degradation. These activities modulate cell, tissue and internal environment stability, and can also affect the occurrence and development of cancer. Previous studies have mostly described autophagy as having dual effects in cancer, serving to limit tumorigenesis in the early stages of cancer, but promoting tumor progression in certain types of cancer. There have been indications in recent years that microRNAs (miRNAs/miRs) and long non‑coding RNAs (lncRNAs), as types of non‑coding RNAs, play major roles in the occurrence, invasion, development and drug resistance of hepatocellular carcinoma (HCC) and in the migration of HCC cells by governing HCC cell autophagy. Therefore, understanding which miRNAs and lncRNAs play such roles and the relevant molecular mechanisms is critical. The present review highlights the significant functions of miRNAs and lncRNAs in the regulation of autophagy in HCC and the relevant mechanisms, aiming to provide novel insight into HCC therapeutics.
Keywords: autophagy; hepatocellular carcinoma; long non‑coding RNAs; microRNAs; non‑coding RNAs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

