Single-cell RNA sequencing reveals the transcriptomic landscape of kidneys in patients with ischemic acute kidney injury
- PMID: 37083129
- PMCID: PMC10278705
- DOI: 10.1097/CM9.0000000000002679
Single-cell RNA sequencing reveals the transcriptomic landscape of kidneys in patients with ischemic acute kidney injury
Abstract
Background: Ischemic acute kidney injury (AKI) is a common syndrome associated with considerable mortality and healthcare costs. Up to now, the underlying pathogenesis of ischemic AKI remains incompletely understood, and specific strategies for early diagnosis and treatment of ischemic AKI are still lacking. Here, this study aimed to define the transcriptomic landscape of AKI patients through single-cell RNA sequencing (scRNA-seq) analysis in kidneys.
Methods: In this study, scRNA-seq technology was applied to kidneys from two ischemic AKI patients, and three human public scRNA-seq datasets were collected as controls. Differentially expressed genes (DEGs) and cell clusters of kidneys were determined. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, as well as the ligand-receptor interaction between cells, were performed. We also validated several DEGs expression in kidneys from human ischemic AKI and ischemia/reperfusion (I/R) injury induced AKI mice through immunohistochemistry staining.
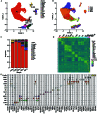
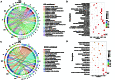
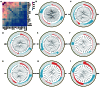
Results: 15 distinct cell clusters were determined in kidney from subjects of ischemic AKI and control. The injured proximal tubules (PT) displayed a proapoptotic and proinflammatory phenotype. PT cells of ischemic AKI had up-regulation of novel pro-apoptotic genes including USP47 , RASSF4 , EBAG9 , IER3 , SASH1 , SEPTIN7 , and NUB1 , which have not been reported in ischemic AKI previously. Several hub genes were validated in kidneys from human AKI and renal I/R injury mice, respectively. Furthermore, PT highly expressed DEGs enriched in endoplasmic reticulum stress, autophagy, and retinoic acid-inducible gene I (RIG-I) signaling. DEGs overexpressed in other tubular cells were primarily enriched in nucleotide-binding and oligomerization domain (NOD)-like receptor signaling, estrogen signaling, interleukin (IL)-12 signaling, and IL-17 signaling. Overexpressed genes in kidney-resident immune cells including macrophages, natural killer T (NKT) cells, monocytes, and dendritic cells were associated with leukocyte activation, chemotaxis, cell adhesion, and complement activation. In addition, the ligand-receptor interactions analysis revealed prominent communications between macrophages and monocytes with other cells in the process of ischemic AKI.
Conclusion: Together, this study reveals distinct cell-specific transcriptomic atlas of kidney in ischemic AKI patients, altered signaling pathways, and potential cell-cell crosstalk in the development of AKI. These data reveal new insights into the pathogenesis and potential therapeutic strategies in ischemic AKI.
Copyright © 2023 The Chinese Medical Association, produced by Wolters Kluwer, Inc. under the CC-BY-NC-ND license.
Conflict of interest statement
None.
Figures

References
-
- Khwaja A. KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin Pract 2012;120: c179–c184. doi: 10.1159/000339789. - PubMed
-
- Mehta RL Cerda J Burdmann EA Tonelli M Garcia-Garcia G Jha V, et al. International Society of Nephrology's 0by25 initiative for acute kidney injury (zero preventable deaths by 2025): A human rights case for nephrology. Lancet 2015;385: 2616–2643. doi: 10.1016/S0140-6736(15)60126-X. - PubMed
-
- Hoste EAJ Kellum JA Selby NM Zarbock A Palevsky PM Bagshaw SM, et al. Global epidemiology and outcomes of acute kidney injury. Nat Rev Nephrol 2018;14: 607–625. doi: 10.1038/s41581-018-0052-0. - PubMed
-
- Hoste EA Bagshaw SM Bellomo R Cely CM Colman R Cruz DN, et al. Epidemiology of acute kidney injury in critically ill patients: The multinational AKI-EPI study. Intensive Care Med 2015;41: 1411–1423. doi: 10.1007/s00134-015-3934-7. - PubMed
-
- Ronco C, Bellomo R, Kellum JA. Acute kidney injury. Lancet 2019;394: 1949–1964. doi: 10.1016/S0140-6736(19)32563-2. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

