The structure of a 12-segmented dsRNA reovirus: New insights into capsid stabilization and organization
- PMID: 37083840
- PMCID: PMC10155992
- DOI: 10.1371/journal.ppat.1011341
The structure of a 12-segmented dsRNA reovirus: New insights into capsid stabilization and organization
Abstract
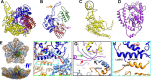
Infecting a wide range of hosts, members of Reovirales (formerly Reoviridae) consist of a genome with different numbers of segmented double stranded RNAs (dsRNA) encapsulated by a proteinaceous shell and carry out genome replication and transcription inside the virion. Several cryo-electron microscopy (cryo-EM) structures of reoviruses with 9, 10 or 11 segmented dsRNA genomes have revealed insights into genome arrangement and transcription. However, the structure and genome arrangement of 12-segmented Reovirales members remain poorly understood. Using cryo-EM, we determined the structure of mud crab reovirus (MCRV), a 12-segmented dsRNA virus that is a putative member of Reovirales in the non-turreted Sedoreoviridae family, to near-atomic resolutions with icosahedral symmetry (3.1 Å) and without imposing icosahedral symmetry (3.4 Å). These structures revealed the organization of the major capsid proteins in two layers: an outer T = 13 layer consisting of VP12 trimers and unique VP11 clamps, and an inner T = 1 layer consisting of VP3 dimers. Additionally, ten RNA dependent RNA polymerases (RdRp) were well resolved just below the VP3 layer but were offset from the 5-fold axes and arranged with D5 symmetry, which has not previously been seen in other members of Reovirales. The N-termini of VP3 were shown to adopt four unique conformations; two of which anchor the RdRps, while the other two conformations are likely involved in genome organization and capsid stability. Taken together, these structures provide a new level of understanding for capsid stabilization and genome organization of segmented dsRNA viruses.
Copyright: © 2023 Zhang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




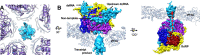
Similar articles
-
In Situ Structures of the Polymerase Complex and RNA Genome Show How Aquareovirus Transcription Machineries Respond to Uncoating.J Virol. 2018 Oct 12;92(21):e00774-18. doi: 10.1128/JVI.00774-18. Print 2018 Nov 1. J Virol. 2018. PMID: 30068643 Free PMC article.
-
Viral Capsid and Polymerase in Reoviridae.Subcell Biochem. 2022;99:525-552. doi: 10.1007/978-3-031-00793-4_17. Subcell Biochem. 2022. PMID: 36151388
-
Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly.Proc Natl Acad Sci U S A. 2018 Jul 10;115(28):7344-7349. doi: 10.1073/pnas.1803885115. Epub 2018 Jun 25. Proc Natl Acad Sci U S A. 2018. PMID: 29941585 Free PMC article.
-
High-resolution 3D structures reveal the biological functions of reoviruses.Virol Sin. 2013 Dec;28(6):318-25. doi: 10.1007/s12250-013-3341-6. Epub 2013 Nov 6. Virol Sin. 2013. PMID: 24254888 Free PMC article. Review.
-
Chrysovirus structure: repeated helical core as evidence of gene duplication.Adv Virus Res. 2013;86:87-108. doi: 10.1016/B978-0-12-394315-6.00004-0. Adv Virus Res. 2013. PMID: 23498904 Review.
Cited by
-
Predictive modeling and cryo-EM: A synergistic approach to modeling macromolecular structure.Biophys J. 2024 Feb 20;123(4):435-450. doi: 10.1016/j.bpj.2024.01.021. Epub 2024 Jan 23. Biophys J. 2024. PMID: 38268190 Free PMC article. Review.
-
Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.Nat Commun. 2024 Mar 13;15(1):2284. doi: 10.1038/s41467-024-46624-x. Nat Commun. 2024. PMID: 38480794 Free PMC article.
-
Characterization of the Virome Associated with the Ubiquitous Two-Spotted Spider Mite, Tetranychus urticae.Viruses. 2024 Sep 27;16(10):1532. doi: 10.3390/v16101532. Viruses. 2024. PMID: 39459865 Free PMC article.
References
-
- Attoui H M P.P.C., Becnel J., Belaganahalli S., Bergoin M., Brussard C.P., Chappell J.D. Ciarlet M., del Vas M., & other authors. Family Reoviridae. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses: Elsevier; 2012.
-
- Dunn SE, Li H, Cardone G, Nibert ML, Ghabrial SA, Baker TS. Three-dimensional structure of victorivirus HvV190S suggests coat proteins in most totiviruses share a conserved core. PLoS pathogens. 2013;9(3):e1003225. Epub 2013/03/22. doi: 10.1371/journal.ppat.1003225 ; PubMed Central PMCID: PMC3597494. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

