MicroRNA-708 emerges as a potential candidate to target undruggable NRAS
- PMID: 37083947
- PMCID: PMC10120925
- DOI: 10.1371/journal.pone.0284744
MicroRNA-708 emerges as a potential candidate to target undruggable NRAS
Abstract
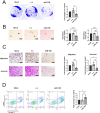
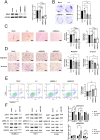
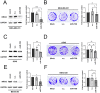
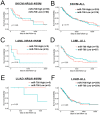
RAS, the most frequently mutated oncogene that drives tumorigenesis by promoting cell proliferation, survival, and motility, has been perceived as undruggable for the past three decades. However, intense research in the past has mainly focused on KRAS mutations, and targeted therapy for NRAS mutations remains an unmet medical need. NRAS mutation is frequently observed in several cancer types, including melanoma (15-20%), leukemia (10%), and occasionally other cancer types. Here, we report using miRNA-708, which targets the distinct 3' untranslated region (3'UTR) of NRAS, to develop miRNA-based precision medicine to treat NRAS mutation-driven cancers. We first confirmed that NRAS is a direct target of miRNA-708. Overexpression of miRNA-708 successfully reduced NRAS protein levels in melanoma, leukemia, and lung cancer cell lines with NRAS mutations, resulting in suppressed cell proliferation, anchorage-independent growth, and promotion of reactive oxygen species-induced apoptosis. Consistent with the functional data, the activities of NRAS-downstream effectors, the PI3K-AKT-mTOR or RAF-MEK-ERK signaling pathway, were impaired in miR-708 overexpressing cells. On the other hand, cell proliferation was not disturbed by miRNA-708 in cell lines carrying wild-type NRAS. Collectively, our data unveil the therapeutic potential of using miRNA-708 in NRAS mutation-driven cancers through direct depletion of constitutively active NRAS and thus inhibition of its downstream effectors to decelerate cancer progression. Harnessing the beneficial effects of miR-708 may therefore offer a potential avenue for small RNA-mediated precision medicine in cancer treatment.
Copyright: © 2023 Pang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

