Single-nucleus transcriptomics reveals a gatekeeper role for FOXP1 in primate cardiac aging
- PMID: 37084237
- PMCID: PMC10120965
- DOI: 10.1093/procel/pwac038
Single-nucleus transcriptomics reveals a gatekeeper role for FOXP1 in primate cardiac aging
Abstract
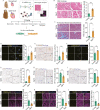
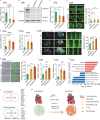
Aging poses a major risk factor for cardiovascular diseases, the leading cause of death in the aged population. However, the cell type-specific changes underlying cardiac aging are far from being clear. Here, we performed single-nucleus RNA-sequencing analysis of left ventricles from young and aged cynomolgus monkeys to define cell composition changes and transcriptomic alterations across different cell types associated with age. We found that aged cardiomyocytes underwent a dramatic loss in cell numbers and profound fluctuations in transcriptional profiles. Via transcription regulatory network analysis, we identified FOXP1, a core transcription factor in organ development, as a key downregulated factor in aged cardiomyocytes, concomitant with the dysregulation of FOXP1 target genes associated with heart function and cardiac diseases. Consistently, the deficiency of FOXP1 led to hypertrophic and senescent phenotypes in human embryonic stem cell-derived cardiomyocytes. Altogether, our findings depict the cellular and molecular landscape of ventricular aging at the single-cell resolution, and identify drivers for primate cardiac aging and potential targets for intervention against cardiac aging and associated diseases.
Keywords: FOXP1; aging; cardiomyocyte; primate; single-nucleus RNA-sequencing.
©The Author(s) 2022. Published by Oxford University Press on behalf of Higher Education Press.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Abdellatif M, Sedej S, Carmona-Gutierrez Det al. . Autophagy in cardiovascular aging. Circ Res 2018;123:803–824. - PubMed
-
- Abplanalp WT, John D, Cremer Set al. . Single-cell RNA-sequencing reveals profound changes in circulating immune cells in patients with heart failure. Cardiovasc Res 2021;117:484–494. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

