Deciphering associations between gut microbiota and clinical factors using microbial modules
- PMID: 37084255
- PMCID: PMC10191612
- DOI: 10.1093/bioinformatics/btad213
Deciphering associations between gut microbiota and clinical factors using microbial modules
Abstract
Motivation: Human gut microbiota plays a vital role in maintaining body health. The dysbiosis of gut microbiota is associated with a variety of diseases. It is critical to uncover the associations between gut microbiota and disease states as well as other intrinsic or environmental factors. However, inferring alterations of individual microbial taxa based on relative abundance data likely leads to false associations and conflicting discoveries in different studies. Moreover, the effects of underlying factors and microbe-microbe interactions could lead to the alteration of larger sets of taxa. It might be more robust to investigate gut microbiota using groups of related taxa instead of the composition of individual taxa.
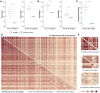
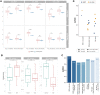
Results: We proposed a novel method to identify underlying microbial modules, i.e. groups of taxa with similar abundance patterns affected by a common latent factor, from longitudinal gut microbiota and applied it to inflammatory bowel disease (IBD). The identified modules demonstrated closer intragroup relationships, indicating potential microbe-microbe interactions and influences of underlying factors. Associations between the modules and several clinical factors were investigated, especially disease states. The IBD-associated modules performed better in stratifying the subjects compared with the relative abundance of individual taxa. The modules were further validated in external cohorts, demonstrating the efficacy of the proposed method in identifying general and robust microbial modules. The study reveals the benefit of considering the ecological effects in gut microbiota analysis and the great promise of linking clinical factors with underlying microbial modules.
Availability and implementation: https://github.com/rwang-z/microbial_module.git.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures




Similar articles
-
The Gut Microbiome and Inflammatory Bowel Diseases.Annu Rev Med. 2022 Jan 27;73:455-468. doi: 10.1146/annurev-med-042320-021020. Epub 2021 Sep 23. Annu Rev Med. 2022. PMID: 34555295 Free PMC article. Review.
-
The Roles of Inflammation, Nutrient Availability and the Commensal Microbiota in Enteric Pathogen Infection.Microbiol Spectr. 2015 Jun;3(3). doi: 10.1128/microbiolspec.MBP-0008-2014. Microbiol Spectr. 2015. PMID: 26185088
-
Interactions and Stability of Gut Microbiota in Zebrafish Increase with Host Development.Microbiol Spectr. 2022 Apr 27;10(2):e0169621. doi: 10.1128/spectrum.01696-21. Epub 2022 Mar 21. Microbiol Spectr. 2022. PMID: 35311546 Free PMC article.
-
Adjusting for age improves identification of gut microbiome alterations in multiple diseases.Elife. 2020 Mar 11;9:e50240. doi: 10.7554/eLife.50240. Elife. 2020. PMID: 32159510 Free PMC article.
-
Gut Microbiota Dysbiosis as Risk and Premorbid Factors of IBD and IBS Along the Childhood-Adulthood Transition.Inflamm Bowel Dis. 2016 Feb;22(2):487-504. doi: 10.1097/MIB.0000000000000602. Inflamm Bowel Dis. 2016. PMID: 26588090 Review.
Cited by
-
Working together: gut microbe-microbe interactions shape host inflammation.Infect Immun. 2025 Jul 8;93(7):e0051224. doi: 10.1128/iai.00512-24. Epub 2025 Jun 13. Infect Immun. 2025. PMID: 40512008 Free PMC article. Review.
-
Disrupted microbial cross-feeding and altered L-phenylalanine consumption in people living with HIV.Brief Bioinform. 2025 Mar 4;26(2):bbaf111. doi: 10.1093/bib/bbaf111. Brief Bioinform. 2025. PMID: 40072847 Free PMC article.
-
Co-expression module analysis reveals high expression homogeneity for both coding and non-coding genes in sepsis.BMC Genomics. 2023 Jul 24;24(1):418. doi: 10.1186/s12864-023-09460-9. BMC Genomics. 2023. PMID: 37488493 Free PMC article.

