Inhibition of MYC translation through targeting of the newly identified PHB-eIF4F complex as a therapeutic strategy in CLL
- PMID: 37084385
- PMCID: PMC10646824
- DOI: 10.1182/blood.2022017839
Inhibition of MYC translation through targeting of the newly identified PHB-eIF4F complex as a therapeutic strategy in CLL
Abstract
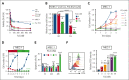
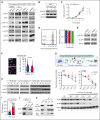
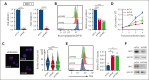
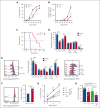
Dysregulation of messenger RNA (mRNA) translation, including preferential translation of mRNA with complex 5' untranslated regions such as the MYC oncogene, is recognized as an important mechanism in cancer. Here, we show that both human and murine chronic lymphocytic leukemia (CLL) cells display a high translation rate, which is inhibited by the synthetic flavagline FL3, a prohibitin (PHB)-binding drug. A multiomics analysis performed in samples from patients with CLL and cell lines treated with FL3 revealed the decreased translation of the MYC oncogene and of proteins involved in cell cycle and metabolism. Furthermore, inhibiting translation induced a proliferation arrest and a rewiring of MYC-driven metabolism. Interestingly, contrary to other models, the RAS-RAF-(PHBs)-MAPK pathway is neither impaired by FL3 nor implicated in translation regulation in CLL cells. Here, we rather show that PHBs are directly associated with the eukaryotic initiation factor (eIF)4F translation complex and are targeted by FL3. Knockdown of PHBs resembled FL3 treatment. Importantly, inhibition of translation controlled CLL development in vivo, either alone or combined with immunotherapy. Finally, high expression of translation initiation-related genes and PHBs genes correlated with poor survival and unfavorable clinical parameters in patients with CLL. Overall, we demonstrated that translation inhibition is a valuable strategy to control CLL development by blocking the translation of several oncogenic pathways including MYC. We also unraveled a new and direct role of PHBs in translation initiation, thus creating new therapeutic opportunities for patients with CLL.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures








Comment in
-
Targeting mRNA translation in CLL.Blood. 2023 Jun 29;141(26):3129-3130. doi: 10.1182/blood.2023020420. Blood. 2023. PMID: 37383005 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

