Genetic subdivisions of follicular lymphoma defined by distinct coding and noncoding mutation patterns
- PMID: 37084389
- PMCID: PMC10644066
- DOI: 10.1182/blood.2022018719
Genetic subdivisions of follicular lymphoma defined by distinct coding and noncoding mutation patterns
Abstract
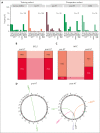
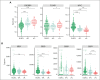
Follicular lymphoma (FL) accounts for ∼20% of all new lymphoma cases. Increases in cytological grade are a feature of the clinical progression of this malignancy, and eventual histologic transformation (HT) to the aggressive diffuse large B-cell lymphoma (DLBCL) occurs in up to 15% of patients. Clinical or genetic features to predict the risk and timing of HT have not been described comprehensively. In this study, we analyzed whole-genome sequencing data from 423 patients to compare the protein coding and noncoding mutation landscapes of untransformed FL, transformed FL, and de novo DLBCL. This revealed 2 genetically distinct subgroups of FL, which we have named DLBCL-like (dFL) and constrained FL (cFL). Each subgroup has distinguishing mutational patterns, aberrant somatic hypermutation rates, and biological and clinical characteristics. We implemented a machine learning-derived classification approach to stratify patients with FL into cFL and dFL subgroups based on their genomic features. Using separate validation cohorts, we demonstrate that cFL status, whether assigned with this full classifier or a single-gene approximation, is associated with a reduced rate of HT. This implies distinct biological features of cFL that constrain its evolution, and we highlight the potential for this classification to predict HT from genetic features present at diagnosis.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: R.D.M. and D.W.S. are named inventors on a patent application describing the double-hit signature. The remaining authors declare no competing financial interests.
Figures





Comment in
-
An AID to follicular lymphoma transformation.Blood. 2023 Aug 10;142(6):500-502. doi: 10.1182/blood.2023020811. Blood. 2023. PMID: 37561540 Free PMC article. No abstract available.
References
-
- Kuruvilla J, Assouline S, Hodgson D, et al. A Canadian evidence-based guideline for the first-line treatment of follicular lymphoma: Joint Consensus of the Lymphoma Canada Scientific Advisory Board. Clin Lymphoma, Myeloma & Leukemia. 2015;15(2):59–74. - PubMed
-
- Swerdlow SH, Campo E, Harris NL, et al. International agency for research on cancer; 2008. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues.
-
- Carbone A, Roulland S, Gloghini A, et al. Follicular lymphoma. Nat Rev Dis Primers. 2019;5(1):83. - PubMed
-
- Miranda-Filho A, Piñeros M, Znaor A, Marcos-Gragera R, Steliarova-Foucher E, Bray F. Global patterns and trends in the incidence of non-Hodgkin lymphoma. Cancer Causes Control. 2019;30(5):489–499. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

