m6 A promotes planarian regeneration
- PMID: 37084418
- PMCID: PMC10212710
- DOI: 10.1111/cpr.13481
m6 A promotes planarian regeneration
Abstract
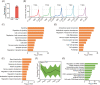
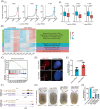
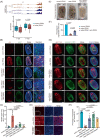
Regeneration is the regrowth of damaged tissues or organs, a vital process in response to damages from primitive organisms to higher mammals. Planarian possesses active whole-body regenerative capability owing to its vast reservoir of adult stem cells, neoblasts, providing an ideal model to delineate the underlying mechanisms for regeneration. RNA N6 -methyladenosine (m6 A) modification participates in many biological processes, including stem cell self-renewal and differentiation, in particular the regeneration of haematopoietic stem cells and axons. However, how m6 A controls regeneration at the whole-organism level remains largely unknown. Here, we demonstrate that the depletion of m6 A methyltransferase regulatory subunit wtap abolishes planarian regeneration, potentially through regulating genes related to cell-cell communication and cell cycle. Single-cell RNA-seq (scRNA-seq) analysis unveils that the wtap knockdown induces a unique type of neural progenitor-like cells (NP-like cells), characterized by specific expression of the cell-cell communication ligand grn. Intriguingly, the depletion of m6 A-modified transcripts grn, cdk9 or cdk7 partially rescues the defective regeneration of planarian caused by wtap knockdown. Overall, our study reveals an indispensable role of m6 A modification in regulating whole-organism regeneration.
© 2023 The Authors. Cell Proliferation published by Beijing Institute for Stem Cell and Regenerative Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Gierer A, Berking S, Bode H, et al. Regeneration of hydra from reaggregated cells. Nat New Biol. 1972;239:98‐101. - PubMed
-
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188‐2190. - PubMed
-
- Wells JM, Watt FM. Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature. 2018;557:322‐328. - PubMed
-
- Alvarado AS, Tsonis PA. Bridging the regeneration gap: genetic insights from diverse animal models. Nat Rev Genet. 2006;7:873‐884. - PubMed
MeSH terms
Grants and funding
- Z201100006820104/Beijing Nova Program
- 20220484210/Beijing Nova Program
- YSBR-073/CAS Project for Young Scientists in Basic Research
- 2018YFA0801200/National Key R&D Program of China
- 91940304/National Natural Science Foundation of China
- 32121001/National Natural Science Foundation of China
- 31770872/National Natural Science Foundation of China
- 32070828/National Natural Science Foundation of China
- 31922017/National Natural Science Foundation of China
- 2017SHZDZX01/Shanghai Municipal Science and Technology Major Project
- XDA16010501/Strategic Priority Research Program of the Chinese Academy of Sciences, China
- XDA16010108/Strategic Priority Research Program of the Chinese Academy of Sciences, China
- Y2022040/Youth Innovation Promotion Association of CAS
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

