The pathogenetic influence of smoking on SARS-CoV-2 infection: Integrative transcriptome and regulomics analysis of lung epithelial cells
- PMID: 37084641
- PMCID: PMC10065815
- DOI: 10.1016/j.compbiomed.2023.106885
The pathogenetic influence of smoking on SARS-CoV-2 infection: Integrative transcriptome and regulomics analysis of lung epithelial cells
Abstract
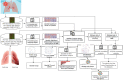
Corona virus disease (COVID-19) has been emerged as pandemic infectious disease. The recent epidemiological data suggest that the smokers are more vulnerable to infection with COVID-19; however, the influence of smoking (SMK) on the COVID-19 infected patients and the mortality is not known yet. In this study, we aimed to discern the influence of SMK on COVID-19 infected patients utilizing the transcriptomics data of COVID-19 infected lung epithelial cells and transcriptomics data smoking matched with controls from lung epithelial cells. The bioinformatics based analysis revealed the molecular insights into the level of transcriptional changes and pathways which are important to identify the impact of smoking on COVID-19 infection and prevalence. We compared differentially expressed genes (DEGs) between COVID-19 and SMK and 59 DEGs were identified as consistently dysregulated at transcriptomics levels. The correlation network analyses were constructed for these common genes using WGCNA R package to see the relationship among these genes. Integration of DEGs with network analysis (protein-protein interaction) showed the presence of 9 hub proteins as key so called "candidate hub proteins" overlapped between COVID-19 patients and SMK. The Gene Ontology and pathways analysis demonstrated the enrichment of inflammatory pathway such as IL-17 signaling pathway, Interleukin-6 signaling, TNF signaling pathway and MAPK1/MAPK3 signaling pathways that might be the therapeutic targets in COVID-19 for smoking persons. The identified genes, pathways, hubs genes, and their regulators might be considered for establishment of key genes and drug targets for SMK and COVID-19.
Keywords: COVID-19; Comorbidity; Drugs; Pathway; Protein–protein interaction; SMOKING.
Copyright © 2023 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

