TRAP1 S-nitrosylation as a model of population-shift mechanism to study the effects of nitric oxide on redox-sensitive oncoproteins
- PMID: 37085483
- PMCID: PMC10121659
- DOI: 10.1038/s41419-023-05780-6
TRAP1 S-nitrosylation as a model of population-shift mechanism to study the effects of nitric oxide on redox-sensitive oncoproteins
Abstract
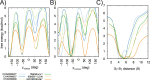
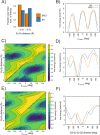
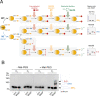
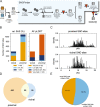
S-nitrosylation is a post-translational modification in which nitric oxide (NO) binds to the thiol group of cysteine, generating an S-nitrosothiol (SNO) adduct. S-nitrosylation has different physiological roles, and its alteration has also been linked to a growing list of pathologies, including cancer. SNO can affect the function and stability of different proteins, such as the mitochondrial chaperone TRAP1. Interestingly, the SNO site (C501) of TRAP1 is in the proximity of another cysteine (C527). This feature suggests that the S-nitrosylated C501 could engage in a disulfide bridge with C527 in TRAP1, resembling the well-known ability of S-nitrosylated cysteines to resolve in disulfide bridge with vicinal cysteines. We used enhanced sampling simulations and in-vitro biochemical assays to address the structural mechanisms induced by TRAP1 S-nitrosylation. We showed that the SNO site induces conformational changes in the proximal cysteine and favors conformations suitable for disulfide bridge formation. We explored 4172 known S-nitrosylated proteins using high-throughput structural analyses. Furthermore, we used a coarse-grained model for 44 protein targets to account for protein flexibility. This resulted in the identification of up to 1248 proximal cysteines, which could sense the redox state of the SNO site, opening new perspectives on the biological effects of redox switches. In addition, we devised two bioinformatic workflows ( https://github.com/ELELAB/SNO_investigation_pipelines ) to identify proximal or vicinal cysteines for a SNO site with accompanying structural annotations. Finally, we analyzed mutations in tumor suppressors or oncogenes in connection with the conformational switch induced by S-nitrosylation. We classified the variants as neutral, stabilizing, or destabilizing for the propensity to be S-nitrosylated and undergo the population-shift mechanism. The methods applied here provide a comprehensive toolkit for future high-throughput studies of new protein candidates, variant classification, and a rich data source for the research community in the NO field.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Identification of new targets of S-nitrosylation in neural stem cells by thiol redox proteomics.Redox Biol. 2020 May;32:101457. doi: 10.1016/j.redox.2020.101457. Epub 2020 Feb 7. Redox Biol. 2020. PMID: 32088623 Free PMC article.
-
Identification of endogenously S-nitrosylated proteins in Arabidopsis plantlets: effect of cold stress on cysteine nitrosylation level.Plant Sci. 2014 Feb;215-216:150-6. doi: 10.1016/j.plantsci.2013.10.014. Epub 2013 Nov 1. Plant Sci. 2014. PMID: 24388526
-
Identification and quantification of S-nitrosylation by cysteine reactive tandem mass tag switch assay.Mol Cell Proteomics. 2012 Feb;11(2):M111.013441. doi: 10.1074/mcp.M111.013441. Epub 2011 Nov 29. Mol Cell Proteomics. 2012. PMID: 22126794 Free PMC article.
-
Protein S-Nitrosylation: Determinants of Specificity and Enzymatic Regulation of S-Nitrosothiol-Based Signaling.Antioxid Redox Signal. 2019 Apr 1;30(10):1331-1351. doi: 10.1089/ars.2017.7403. Epub 2018 Jan 10. Antioxid Redox Signal. 2019. PMID: 29130312 Free PMC article. Review.
-
Screening systems for the identification of S-nitrosylated proteins.Nitric Oxide. 2011 Aug 1;25(2):108-11. doi: 10.1016/j.niox.2010.11.002. Epub 2010 Nov 24. Nitric Oxide. 2011. PMID: 21111056 Review.
Cited by
-
The evolution of S-nitrosylation detection methodology and the role of protein S-nitrosylation in various cancers.Cancer Cell Int. 2024 Dec 19;24(1):408. doi: 10.1186/s12935-024-03568-y. Cancer Cell Int. 2024. PMID: 39702281 Free PMC article. Review.
-
Exploring protein functions from structural flexibility using CABS-flex modeling.Protein Sci. 2024 Sep;33(9):e5090. doi: 10.1002/pro.5090. Protein Sci. 2024. PMID: 39194135 Free PMC article. Review.
-
Protein Oxidative Modifications in Neurodegenerative Diseases: From Advances in Detection and Modelling to Their Use as Disease Biomarkers.Antioxidants (Basel). 2024 May 31;13(6):681. doi: 10.3390/antiox13060681. Antioxidants (Basel). 2024. PMID: 38929122 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

