Control of a hippocampal recurrent excitatory circuit by cannabinoid receptor-interacting protein Gap43
- PMID: 37085487
- PMCID: PMC10121561
- DOI: 10.1038/s41467-023-38026-2
Control of a hippocampal recurrent excitatory circuit by cannabinoid receptor-interacting protein Gap43
Abstract
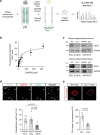
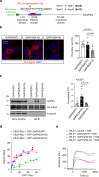
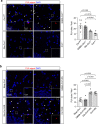
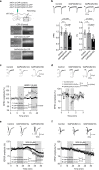
The type-1 cannabinoid receptor (CB1R) is widely expressed in excitatory and inhibitory nerve terminals, and by suppressing neurotransmitter release, its activation modulates neural circuits and brain function. While the interaction of CB1R with various intracellular proteins is thought to alter receptor signaling, the identity and role of these proteins are poorly understood. Using a high-throughput proteomic analysis complemented with an array of in vitro and in vivo approaches in the mouse brain, we report that the C-terminal, intracellular domain of CB1R interacts specifically with growth-associated protein of 43 kDa (GAP43). The CB1R-GAP43 interaction occurs selectively at mossy cell axon boutons, which establish excitatory synapses with dentate granule cells in the hippocampus. This interaction impairs CB1R-mediated suppression of mossy cell to granule cell transmission, thereby inhibiting cannabinoid-mediated anti-convulsant activity in mice. Thus, GAP43 acts as a synapse type-specific regulatory partner of CB1R that hampers CB1R-mediated effects on hippocampal circuit function.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

