Transcription tuned by S-nitrosylation underlies a mechanism for Staphylococcus aureus to circumvent vancomycin killing
- PMID: 37085493
- PMCID: PMC10120478
- DOI: 10.1038/s41467-023-37949-0
Transcription tuned by S-nitrosylation underlies a mechanism for Staphylococcus aureus to circumvent vancomycin killing
Erratum in
-
Author Correction: Transcription tuned by S-nitrosylation underlies a mechanism for Staphylococcus aureus to circumvent vancomycin killing.Nat Commun. 2023 Jun 23;14(1):3748. doi: 10.1038/s41467-023-39231-9. Nat Commun. 2023. PMID: 37353476 Free PMC article. No abstract available.
Abstract
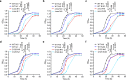
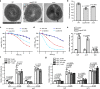
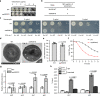
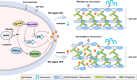
Treatment of Staphylococcus aureus infections is a constant challenge due to emerging resistance to vancomycin, a last-resort drug. S-nitrosylation, the covalent attachment of a nitric oxide (NO) group to a cysteine thiol, mediates redox-based signaling for eukaryotic cellular functions. However, its role in bacteria is largely unknown. Here, proteomic analysis revealed that S-nitrosylation is a prominent growth feature of vancomycin-intermediate S. aureus. Deletion of NO synthase (NOS) or removal of S-nitrosylation from the redox-sensitive regulator MgrA or WalR resulted in thinner cell walls and increased vancomycin susceptibility, which was due to attenuated promoter binding and released repression of genes involved in cell wall metabolism. These genes failed to respond to H2O2-induced oxidation, suggesting distinct transcriptional responses to alternative modifications of the cysteine residue. Furthermore, treatment with a NOS inhibitor significantly decreased vancomycin resistance in S. aureus. This study reveals that transcriptional regulation via S-nitrosylation underlies a mechanism for NO-mediated bacterial antibiotic resistance.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Cervimycin-Resistant Staphylococcus aureus Strains Display Vancomycin-Intermediate Resistant Phenotypes.Microbiol Spectr. 2022 Oct 26;10(5):e0256722. doi: 10.1128/spectrum.02567-22. Epub 2022 Sep 29. Microbiol Spectr. 2022. PMID: 36173303 Free PMC article.
-
Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR.PLoS Pathog. 2011 Nov;7(11):e1002359. doi: 10.1371/journal.ppat.1002359. Epub 2011 Nov 10. PLoS Pathog. 2011. PMID: 22102812 Free PMC article.
-
Reversible antibiotic tolerance induced in Staphylococcus aureus by concurrent drug exposure.mBio. 2015 Jan 13;6(1):e02268-14. doi: 10.1128/mBio.02268-14. mBio. 2015. PMID: 25587013 Free PMC article.
-
Mechanisms of vancomycin resistance in Staphylococcus aureus.J Clin Invest. 2014 Jul;124(7):2836-40. doi: 10.1172/JCI68834. Epub 2014 Jul 1. J Clin Invest. 2014. PMID: 24983424 Free PMC article. Review.
-
Clinical implications of vancomycin heteroresistant and intermediately susceptible Staphylococcus aureus.Pharmacotherapy. 2015 Apr;35(4):424-32. doi: 10.1002/phar.1577. Pharmacotherapy. 2015. PMID: 25884530 Review.
Cited by
-
Regulation of bacterial phosphorelay systems.RSC Chem Biol. 2025 Jun 19;6(8):1252-1269. doi: 10.1039/d5cb00016e. eCollection 2025 Jul 30. RSC Chem Biol. 2025. PMID: 40575134 Free PMC article. Review.
-
Alkaline shock protein 23 (Asp23)-controlled cell wall imbalance promotes membrane vesicle biogenesis in Staphylococcus aureus.J Extracell Vesicles. 2024 Sep;13(9):e12501. doi: 10.1002/jev2.12501. J Extracell Vesicles. 2024. PMID: 39193667 Free PMC article.
-
Factors impacting the regulation of nos gene expression in Staphylococcus aureus.Microbiol Spectr. 2023 Sep 25;11(5):e0168823. doi: 10.1128/spectrum.01688-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37747881 Free PMC article.
-
S-Nitrosylation of NOTCH1 Regulates Mesenchymal Stem Cells Differentiation Into Hepatocyte-Like Cells by Inhibiting Notch Signalling Pathway.J Cell Mol Med. 2024 Dec;28(23):e70274. doi: 10.1111/jcmm.70274. J Cell Mol Med. 2024. PMID: 39656437 Free PMC article.
-
Metal natural product complex Ru-procyanidins with quadruple enzymatic activity combat infections from drug-resistant bacteria.Acta Pharm Sin B. 2024 May;14(5):2298-2316. doi: 10.1016/j.apsb.2023.12.017. Epub 2024 Jan 26. Acta Pharm Sin B. 2024. PMID: 38799629 Free PMC article.
References
-
- Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 2010;23:99–139. - PMC - PubMed
-
- Liu J, Gefen O, Ronin I, Bar-Meir M, Balaban NQ. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science. 2020;367:200–204. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

