Transcriptomics unravels molecular changes associated with cilia and COVID-19 in chronic rhinosinusitis with nasal polyps
- PMID: 37085563
- PMCID: PMC10121071
- DOI: 10.1038/s41598-023-32944-3
Transcriptomics unravels molecular changes associated with cilia and COVID-19 in chronic rhinosinusitis with nasal polyps
Abstract
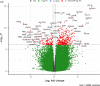
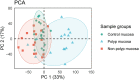
Chronic rhinosinusitis with nasal polyps (CRSwNP) is a common upper respiratory tract complication where the pathogenesis is largely unknown. Herein, we investigated the transcriptome profile in nasal mucosa biopsies of CRSwNP patients and healthy individuals. We further integrated the transcriptomics data with genes located in chromosomal regions containing genome-wide significant gene variants for COVID-19. Among the most significantly upregulated genes in polyp mucosa were CCL18, CLEC4G, CCL13 and SLC9A3. Pathways involving "Ciliated epithelial cells" were the most differentially expressed molecular pathways when polyp mucosa and non-polyp mucosa from the same patient was compared. Natural killer T-cell (NKT) and viral pathways were the most statistically significant pathways in the mucosa of CRSwNP patients compared with those of healthy control individuals. Upregulated genes in polyp mucosa, located within the genome-wide associated regions of COVID-19, included LZTFL1, CCR9, SLC6A20, IFNAR1, IFNAR2 and IL10RB. Interestingly, the second most over-expressed gene in our study, CLEC4G, has been shown to bind directly to SARS-CoV-2 spike's N-terminal domain and mediate its entry and infection. Our results on altered expression of genes related to cilia and viruses point to the de-regulation of viral defenses in CRSwNP patients, and may give clues to future intervention strategies.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

