Mucin-microbiome signatures shape the tumor microenvironment in gastric cancer
- PMID: 37085819
- PMCID: PMC10120190
- DOI: 10.1186/s40168-023-01534-w
Mucin-microbiome signatures shape the tumor microenvironment in gastric cancer
Abstract
Background and aims: We aimed to identify mucin-microbiome signatures shaping the tumor microenvironment in gastric adenocarcinomas and clinical outcomes.
Methods: We performed high-throughput profiling of the mucin phenotypes present in 108 gastric adenocarcinomas and 20 functional dyspepsia cases using validated mucin-based RT-qPCRs with subsequent immunohistochemistry validation and correlated the data with clinical outcome parameters. The gastric microbiota was assessed by 16S rRNA gene sequencing, taxonomy, and community composition determined, microbial networks analyzed, and the metagenome inferred in association with mucin phenotypes and expression.
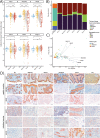
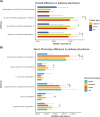
Results: Gastric adenocarcinomas with an intestinal mucin environment or high-level MUC13 expression are associated with poor survival. On the contrary, gastric MUC5AC or MUC6 abundance was associated with a more favorable outcome. The oral taxa Neisseria, Prevotella, and Veillonella had centralities in tumors with intestinal and mixed phenotypes and were associated with MUC13 overexpression, highlighting their role as potential drivers in MUC13 signaling in GC. Furthermore, dense bacterial networks were observed in intestinal and mixed mucin phenotype tumors whereas the lowest community complexity was shown in null mucin phenotype tumors due to higher Helicobacter abundance resulting in a more decreased diversity. Enrichment of oral or intestinal microbes was mucin phenotype dependent. More specifically, intestinal mucin phenotype tumors favored the establishment of pro-inflammatory oral taxa forming strong co-occurrence networks.
Conclusions: Our results emphasize key roles for mucins in gastric cancer prognosis and shaping microbial networks in the tumor microenvironment. Specifically, the enriched oral taxa associated with aberrant MUC13 expression can be potential biomarkers in predicting disease outcomes. Video Abstract.
Keywords: Microbiota; Mucin; Stomach cancer; Survival.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Correa P. Human gastric carcinogenesis: a multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52(24):6735-40. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

