Identification of a sub-group of critically ill patients with high risk of intensive care unit-acquired infections and poor clinical course using a transcriptomic score
- PMID: 37085849
- PMCID: PMC10119529
- DOI: 10.1186/s13054-023-04436-3
Identification of a sub-group of critically ill patients with high risk of intensive care unit-acquired infections and poor clinical course using a transcriptomic score
Abstract
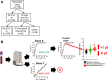
Background: The development of stratification tools based on the assessment of circulating mRNA of genes involved in the immune response is constrained by the heterogeneity of septic patients. The aim of this study is to develop a transcriptomic score based on a pragmatic combination of immune-related genes detected with a prototype multiplex PCR tool.
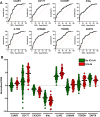
Methods: As training cohort, we used the gene expression dataset obtained from 176 critically ill patients enrolled in the REALISM study (NCT02638779) with various etiologies and still hospitalized in intensive care unit (ICU) at day 5-7. Based on the performances of each gene taken independently to identify patients developing ICU-acquired infections (ICU-AI) after day 5-7, we built an unweighted score assuming the independence of each gene. We then determined the performances of this score to identify a subgroup of patients at high risk to develop ICU-AI, and both longer ICU length of stay and mortality of this high-risk group were assessed. Finally, we validated the effectiveness of this score in a retrospective cohort of 257 septic patients.
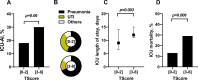
Results: This transcriptomic score (TScore) enabled the identification of a high-risk group of patients (49%) with an increased rate of ICU-AI when compared to the low-risk group (49% vs. 4%, respectively), with longer ICU length of stay (13 days [95% CI 8-30] vs. 7 days [95% CI 6-9], p < 0.001) and higher ICU mortality (15% vs. 2%). High-risk patients exhibited biological features of immune suppression with low monocytic HLA-DR levels, higher immature neutrophils rates and higher IL10 concentrations. Using the TScore, we identified 160 high-risk patients (62%) in the validation cohort, with 30% of ICU-AI (vs. 18% in the low-risk group, p = 0.06), and significantly higher mortality and longer ICU length of stay.
Conclusions: The transcriptomic score provides a useful and reliable companion diagnostic tool to further develop immune modulating drugs in sepsis in the context of personalized medicine.
Keywords: Acquired infections; Intensive care unit; Personalized medicine; Sepsis; Transcriptomic.
© 2023. The Author(s).
Conflict of interest statement
MB, MC, AF, MAC, EC, EP, KBP and JFL are bioMérieux’s employees. JFL and GM are co-inventors in patent applications covering the previous markers. bioFire—a bioMérieux company—holds patents on the technology. This does not alter the authors’ adherence to all the policies on sharing data and materials.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

