Augmin is a Ran-regulated spindle assembly factor
- PMID: 37086784
- PMCID: PMC10318467
- DOI: 10.1016/j.jbc.2023.104736
Augmin is a Ran-regulated spindle assembly factor
Abstract
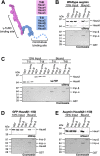
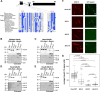
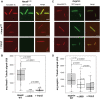
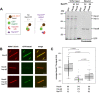
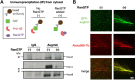
Mitotic spindles are composed of microtubules (MTs) that must nucleate at the right place and time. Ran regulates this process by directly controlling the release of spindle assembly factors (SAFs) from nucleocytoplasmic shuttle proteins importin-αβ and subsequently forms a biochemical gradient of SAFs localized around chromosomes. The majority of spindle MTs are generated by branching MT nucleation, which has been shown to require an eight-subunit protein complex known as augmin. In Xenopus laevis, Ran can control branching through a canonical SAF, TPX2, which is nonessential in Drosophila melanogaster embryos and HeLa cells. Thus, how Ran regulates branching MT nucleation when TPX2 is not required remains unknown. Here, we use in vitro pulldowns and total internal reflection fluorescence microscopy to show that augmin is a Ran-regulated SAF. We demonstrate that augmin directly interacts with both importin-α and importin-β through two nuclear localization sequences on the Haus8 subunit, which overlap with the MT-binding site. Moreover, we show that Ran controls localization of augmin to MTs in both Xenopus egg extract and in vitro. Our results demonstrate that RanGTP directly regulates augmin, which establishes a new way by which Ran controls branching MT nucleation and spindle assembly both in the absence and presence of TPX2.
Keywords: Ran pathway; TPX2; augmin; branching microtubule nucleation; importins; spindle assembly factor.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures
Similar articles
-
Microtubule binding of the human augmin complex is directly controlled by importins and Ran-GTP.J Cell Sci. 2023 Jun 15;136(12):jcs261096. doi: 10.1242/jcs.261096. Epub 2023 Jun 26. J Cell Sci. 2023. PMID: 37357828 Free PMC article.
-
Cdk11 is a RanGTP-dependent microtubule stabilization factor that regulates spindle assembly rate.J Cell Biol. 2008 Mar 10;180(5):867-75. doi: 10.1083/jcb.200706189. Epub 2008 Mar 3. J Cell Biol. 2008. PMID: 18316407 Free PMC article.
-
Microtubule nucleation in mitosis by a RanGTP-dependent protein complex.Curr Biol. 2015 Jan 19;25(2):131-140. doi: 10.1016/j.cub.2014.11.025. Epub 2014 Dec 18. Curr Biol. 2015. PMID: 25532896
-
The mechanism of spindle assembly: functions of Ran and its target TPX2.J Cell Biol. 2004 Sep 27;166(7):949-55. doi: 10.1083/jcb.200312112. J Cell Biol. 2004. PMID: 15452138 Free PMC article. Review.
-
Microtubule nucleation for spindle assembly: one molecule at a time.Trends Biochem Sci. 2023 Sep;48(9):761-775. doi: 10.1016/j.tibs.2023.06.004. Epub 2023 Jul 21. Trends Biochem Sci. 2023. PMID: 37482516 Free PMC article. Review.
Cited by
-
Ran-GTP assembles a specialized spindle structure for accurate chromosome segregation in medaka early embryos.Nat Commun. 2024 Feb 1;15(1):981. doi: 10.1038/s41467-024-45251-w. Nat Commun. 2024. PMID: 38302485 Free PMC article.
-
Roles of Tubulin Concentration during Prometaphase and Ran-GTP during Anaphase of C. elegans meiosis.bioRxiv [Preprint]. 2024 Jun 25:2024.04.19.590357. doi: 10.1101/2024.04.19.590357. bioRxiv. 2024. Update in: Life Sci Alliance. 2024 Jul 3;7(9):e202402884. doi: 10.26508/lsa.202402884. PMID: 38659754 Free PMC article. Updated. Preprint.
-
HURP facilitates spindle assembly by stabilizing microtubules and working synergistically with TPX2.bioRxiv [Preprint]. 2023 Dec 18:2023.12.18.571906. doi: 10.1101/2023.12.18.571906. bioRxiv. 2023. Update in: Nat Commun. 2024 Nov 8;15(1):9689. doi: 10.1038/s41467-024-53630-6. PMID: 38187686 Free PMC article. Updated. Preprint.
-
γ-TuRC asymmetry induces local protofilament mismatch at the RanGTP-stimulated microtubule minus end.EMBO J. 2024 May;43(10):2062-2085. doi: 10.1038/s44318-024-00087-4. Epub 2024 Apr 10. EMBO J. 2024. PMID: 38600243 Free PMC article.
-
HURP facilitates spindle assembly by stabilizing microtubules and working synergistically with TPX2.Nat Commun. 2024 Nov 8;15(1):9689. doi: 10.1038/s41467-024-53630-6. Nat Commun. 2024. PMID: 39516491 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

