Causal feature selection using a knowledge graph combining structured knowledge from the biomedical literature and ontologies: A use case studying depression as a risk factor for Alzheimer's disease
- PMID: 37086959
- PMCID: PMC10355339
- DOI: 10.1016/j.jbi.2023.104368
Causal feature selection using a knowledge graph combining structured knowledge from the biomedical literature and ontologies: A use case studying depression as a risk factor for Alzheimer's disease
Abstract
Background: Causal feature selection is essential for estimating effects from observational data. Identifying confounders is a crucial step in this process. Traditionally, researchers employ content-matter expertise and literature review to identify confounders. Uncontrolled confounding from unidentified confounders threatens validity, conditioning on intermediate variables (mediators) weakens estimates, and conditioning on common effects (colliders) induces bias. Additionally, without special treatment, erroneous conditioning on variables combining roles introduces bias. However, the vast literature is growing exponentially, making it infeasible to assimilate this knowledge. To address these challenges, we introduce a novel knowledge graph (KG) application enabling causal feature selection by combining computable literature-derived knowledge with biomedical ontologies. We present a use case of our approach specifying a causal model for estimating the total causal effect of depression on the risk of developing Alzheimer's disease (AD) from observational data.
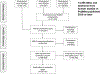
Methods: We extracted computable knowledge from a literature corpus using three machine reading systems and inferred missing knowledge using logical closure operations. Using a KG framework, we mapped the output to target terminologies and combined it with ontology-grounded resources. We translated epidemiological definitions of confounder, collider, and mediator into queries for searching the KG and summarized the roles played by the identified variables. We compared the results with output from a complementary method and published observational studies and examined a selection of confounding and combined role variables in-depth.
Results: Our search identified 128 confounders, including 58 phenotypes, 47 drugs, 35 genes, 23 collider, and 16 mediator phenotypes. However, only 31 of the 58 confounder phenotypes were found to behave exclusively as confounders, while the remaining 27 phenotypes played other roles. Obstructive sleep apnea emerged as a potential novel confounder for depression and AD. Anemia exemplified a variable playing combined roles.
Conclusion: Our findings suggest combining machine reading and KG could augment human expertise for causal feature selection. However, the complexity of causal feature selection for depression with AD highlights the need for standardized field-specific databases of causal variables. Further work is needed to optimize KG search and transform the output for human consumption.
Keywords: Alzheimer’s disease; Causal modeling; Depression; Feature selection; Knowledge graphs; Knowledge representation, management, or engineering.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
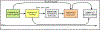
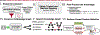







Similar articles
-
Using computable knowledge mined from the literature to elucidate confounders for EHR-based pharmacovigilance.J Biomed Inform. 2021 May;117:103719. doi: 10.1016/j.jbi.2021.103719. Epub 2021 Mar 11. J Biomed Inform. 2021. PMID: 33716168 Free PMC article.
-
Causal Knowledge as a Prerequisite for Interrogating Bias: Reflections on Hernán et al. 20 Years Later.Am J Epidemiol. 2023 Nov 3;192(11):1797-1800. doi: 10.1093/aje/kwab274. Am J Epidemiol. 2023. PMID: 34791035
-
Educational Note: Paradoxical collider effect in the analysis of non-communicable disease epidemiological data: a reproducible illustration and web application.Int J Epidemiol. 2019 Apr 1;48(2):640-653. doi: 10.1093/ije/dyy275. Int J Epidemiol. 2019. PMID: 30561628 Free PMC article.
-
COVID-19 and the epistemology of epidemiological models at the dawn of AI.Ann Hum Biol. 2020 Sep;47(6):506-513. doi: 10.1080/03014460.2020.1839132. Ann Hum Biol. 2020. PMID: 33228409 Review.
-
Using Causal Diagrams to Improve the Design and Interpretation of Medical Research.Chest. 2020 Jul;158(1S):S21-S28. doi: 10.1016/j.chest.2020.03.011. Chest. 2020. PMID: 32658648 Review.
Cited by
-
Knowledge graph and its application in the study of neurological and mental disorders.Front Psychiatry. 2025 Mar 18;16:1452557. doi: 10.3389/fpsyt.2025.1452557. eCollection 2025. Front Psychiatry. 2025. PMID: 40171303 Free PMC article. Review.
-
Development and evaluation of a 4M taxonomy from nursing home staff text messages using a fine-tuned generative language model.J Am Med Inform Assoc. 2025 Mar 1;32(3):535-544. doi: 10.1093/jamia/ocaf006. J Am Med Inform Assoc. 2025. PMID: 39812778 Free PMC article.
-
An open source knowledge graph ecosystem for the life sciences.Sci Data. 2024 Apr 11;11(1):363. doi: 10.1038/s41597-024-03171-w. Sci Data. 2024. PMID: 38605048 Free PMC article.
-
Automated information extraction model enhancing traditional Chinese medicine RCT evidence extraction (Evi-BERT): algorithm development and validation.Front Artif Intell. 2024 Aug 15;7:1454945. doi: 10.3389/frai.2024.1454945. eCollection 2024. Front Artif Intell. 2024. PMID: 39210937 Free PMC article.
-
A Unified Framework for Alzheimer's Disease Knowledge Graphs: Architectures, Principles, and Clinical Translation.Brain Sci. 2025 May 19;15(5):523. doi: 10.3390/brainsci15050523. Brain Sci. 2025. PMID: 40426694 Free PMC article. Review.
References
-
- Cartwright N.Are RCTs the Gold Standard? BioSocieties [Internet]. 2007. Mar [cited 2017 Jul 21];2(1):11–20. Available from: http://www.palgrave-journals.com/doifinder/10.1017/S1745855207005029 - DOI
-
- VanderWeele TJ. Principles of confounder selection. Eur J Epidemiol [Internet]. 2019. [cited 2019 Aug 20];34(3):211–9. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6447501/ - PMC - PubMed
-
- Arntzenius F.Reichenbach’s Common Cause Principle. In: Zalta EN, editor. The Stanford Encyclopedia of Philosophy [Internet]. Fall 2010. Metaphysics Research Lab, Stanford University; 2010 [cited 2019 Dec 10]. p. 1. Available from: https://plato.stanford.edu/archives/fall2010/entries/physics-Rpcc/
-
- VanderWeele TJ, Shpitser I. A new criterion for confounder selection. Biometrics [Internet]. 2011. Dec;67(4):1406–13. Available from: https://www.ncbi.nlm.nih.gov/pubmed/21627630 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

