The hindgut microbiome contributes to host oxidative stress in postpartum dairy cows by affecting glutathione synthesis process
- PMID: 37087457
- PMCID: PMC10122372
- DOI: 10.1186/s40168-023-01535-9
The hindgut microbiome contributes to host oxidative stress in postpartum dairy cows by affecting glutathione synthesis process
Abstract
Background: Dairy cows are susceptible to postpartum systemic oxidative stress (OS), which leads to significant production loss and metabolic disorders. The gut microbiota has been linked to host health and stress levels. However, to what extent the gut microbiota is associated with postpartum OS remains unknown. In this study, the contribution of the fecal microbiota to postpartum systemic OS and its underlying mechanisms were investigated by integrating 16S rRNA gene sequencing, metagenomics, and metabolomics in postpartum dairy cattle and by transplanting fecal microbiota from cattle to mice.
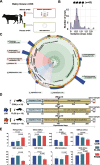
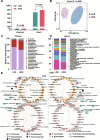
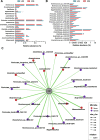
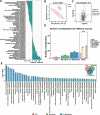
Results: A strong link was found between fecal microbial composition and postpartum OS, with an explainability of 43.1%. A total of 17 significantly differential bacterial genera and 19 species were identified between cows with high (HOS) and low OS (LOS). Among them, 9 genera and 16 species showed significant negative correlations with OS, and Marasmitruncus and Ruminococcus_sp._CAG:724 had the strongest correlations. The microbial functional analysis showed that the fecal microbial metabolism of glutamine, glutamate, glycine, and cysteine involved in glutathione synthesis was lower in HOS cows. Moreover, 58 significantly different metabolites were identified between HOS and LOS cows, and of these metabolites, 19 were produced from microbiota or cometabolism of microbiota and host. Furthermore, these microbial metabolites were enriched in the metabolism of glutamine, glutamate, glycine, and cysteine. The mice gavaged with HOS fecal microbiota had significantly higher OS and lower plasma glutathione peroxidase and glutathione content than those orally administered saline or LOS fecal microbiota.
Conclusions: Integrated results suggest that the fecal microbiota is responsible for OS and that lower glutathione production plays a causative role in HOS. These findings provide novel insights into the mechanisms of postpartum OS and potential regulatory strategies to alleviate OS in dairy cows. Video Abstract.
Keywords: Dairy cows; Fecal microbiota transplantation; Glutathione synthesis; Multiomics; Oxidative stress; Transition period.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Alterations in the gut microbiota and its metabolites contribute to metabolic maladaptation in dairy cows during the development of hyperketonemia.mSystems. 2024 Apr 16;9(4):e0002324. doi: 10.1128/msystems.00023-24. Epub 2024 Mar 19. mSystems. 2024. PMID: 38501812 Free PMC article.
-
Multi-omics profiling of dairy cattle oxidative stress identifies hindgut-derived Phascolarctobacterium succinatutens exhibiting antioxidant activity.NPJ Biofilms Microbiomes. 2025 Apr 22;11(1):61. doi: 10.1038/s41522-025-00698-7. NPJ Biofilms Microbiomes. 2025. PMID: 40263287 Free PMC article.
-
Altered Fecal Microbiome and Correlations of the Metabolome with Plasma Metabolites in Dairy Cows with Left Displaced Abomasum.Microbiol Spectr. 2022 Dec 21;10(6):e0197222. doi: 10.1128/spectrum.01972-22. Epub 2022 Oct 12. Microbiol Spectr. 2022. PMID: 36222683 Free PMC article.
-
Integrated multi-omics analysis reveals the positive leverage of citrus flavonoids on hindgut microbiota and host homeostasis by modulating sphingolipid metabolism in mid-lactation dairy cows consuming a high-starch diet.Microbiome. 2023 Oct 25;11(1):236. doi: 10.1186/s40168-023-01661-4. Microbiome. 2023. PMID: 37880759 Free PMC article.
-
Deterministic succession patterns in the rumen and fecal microbiome associate with host metabolic shifts in peripartum dairy cattle.Gigascience. 2025 Jan 6;14:giaf042. doi: 10.1093/gigascience/giaf042. Gigascience. 2025. PMID: 40388308 Free PMC article.
Cited by
-
Impacts of trehalose supplementation on ruminal microbiota and productivity of Japanese Black heifers under heat-stressed conditions.Anim Biosci. 2025 Jul;38(7):1446-1458. doi: 10.5713/ab.24.0468. Epub 2025 Jan 24. Anim Biosci. 2025. PMID: 39901715 Free PMC article.
-
Melatonin alleviates high temperature exposure induced fetal growth restriction via the gut-placenta-fetus axis in pregnant mice.J Adv Res. 2025 Feb;68:131-146. doi: 10.1016/j.jare.2024.02.014. Epub 2024 Feb 20. J Adv Res. 2025. PMID: 38382594 Free PMC article.
-
Alanine Derived from Ruminococcus_E bovis Alleviates Energy Metabolic Disorders during the Peripartum Period by Providing Glucogenic Precursors.Research (Wash D C). 2025 Apr 25;8:0682. doi: 10.34133/research.0682. eCollection 2025. Research (Wash D C). 2025. PMID: 40290137 Free PMC article.
-
Comparison of lipidome profiles in serum from lactating dairy cows supplemented with Acremonium terrestris culture based on UPLC-QTRAP-MS/MS.BMC Biotechnol. 2024 Aug 12;24(1):56. doi: 10.1186/s12896-024-00881-2. BMC Biotechnol. 2024. PMID: 39135176 Free PMC article.
-
Alterations in the gut microbiota and its metabolites contribute to metabolic maladaptation in dairy cows during the development of hyperketonemia.mSystems. 2024 Apr 16;9(4):e0002324. doi: 10.1128/msystems.00023-24. Epub 2024 Mar 19. mSystems. 2024. PMID: 38501812 Free PMC article.
References
-
- Ingvartsen KL. Feeding and management-related diseases in the transition cow: physiological adaptations around calving and strategies to reduce feeding-related diseases. Anim Feed Sci Technol. 2006;126:175–213. doi: 10.1016/j.anifeedsci.2005.08.003. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

