HaploCoV: unsupervised classification and rapid detection of novel emerging variants of SARS-CoV-2
- PMID: 37087497
- PMCID: PMC10122080
- DOI: 10.1038/s42003-023-04784-4
HaploCoV: unsupervised classification and rapid detection of novel emerging variants of SARS-CoV-2
Abstract
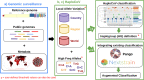
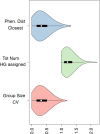
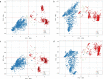
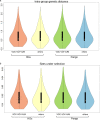
Accurate and timely monitoring of the evolution of SARS-CoV-2 is crucial for identifying and tracking potentially more transmissible/virulent viral variants, and implement mitigation strategies to limit their spread. Here we introduce HaploCoV, a novel software framework that enables the exploration of SARS-CoV-2 genomic diversity through space and time, to identify novel emerging viral variants and prioritize variants of potential epidemiological interest in a rapid and unsupervised manner. HaploCoV can integrate with any classification/nomenclature and incorporates an effective scoring system for the prioritization of SARS-CoV-2 variants. By performing retrospective analyses of more than 11.5 M genome sequences we show that HaploCoV demonstrates high levels of accuracy and reproducibility and identifies the large majority of epidemiologically relevant viral variants - as flagged by international health authorities - automatically and with rapid turn-around times.Our results highlight the importance of the application of strategies based on the systematic analysis and integration of regional data for rapid identification of novel, emerging variants of SARS-CoV-2. We believe that the approach outlined in this study will contribute to relevant advances to current and future genomic surveillance methods.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Early detection of SARS-CoV-2 variants through dynamic co-mutation network surveillance.Front Public Health. 2023 Jan 23;11:1015969. doi: 10.3389/fpubh.2023.1015969. eCollection 2023. Front Public Health. 2023. PMID: 36755900 Free PMC article.
-
Comparative Genomics Reveals Early Emergence and Biased Spatiotemporal Distribution of SARS-CoV-2.Mol Biol Evol. 2021 May 19;38(6):2547-2565. doi: 10.1093/molbev/msab049. Mol Biol Evol. 2021. PMID: 33605421 Free PMC article.
-
Emergency SARS-CoV-2 Variants of Concern: Novel Multiplex Real-Time RT-PCR Assay for Rapid Detection and Surveillance.Microbiol Spectr. 2022 Feb 23;10(1):e0251321. doi: 10.1128/spectrum.02513-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196812 Free PMC article.
-
Evolution of SARS-CoV-2 Variants: Implications on Immune Escape, Vaccination, Therapeutic and Diagnostic Strategies.Viruses. 2023 Apr 10;15(4):944. doi: 10.3390/v15040944. Viruses. 2023. PMID: 37112923 Free PMC article. Review.
-
Genomic surveillance to combat COVID-19: challenges and opportunities.Lancet Microbe. 2021 Sep;2(9):e481-e484. doi: 10.1016/S2666-5247(21)00121-X. Epub 2021 Jul 27. Lancet Microbe. 2021. PMID: 34337584 Free PMC article. Review.
Cited by
-
Data-driven recombination detection in viral genomes.Nat Commun. 2024 Apr 17;15(1):3313. doi: 10.1038/s41467-024-47464-5. Nat Commun. 2024. PMID: 38632281 Free PMC article.
-
Rapid SARS-CoV-2 surveillance using clinical, pooled, or wastewater sequence as a sensor for population change.Genome Res. 2024 Oct 29;34(10):1651-1660. doi: 10.1101/gr.278594.123. Genome Res. 2024. PMID: 39322283 Free PMC article.
-
A systematic review of hypervirulent Klebsiella pneumoniae research: bibliometric and topic modeling perspectives.Front Med (Lausanne). 2025 Apr 11;12:1545678. doi: 10.3389/fmed.2025.1545678. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40291031 Free PMC article.
-
mapPat: tracking pathogens evolution in space and time.Bioinform Adv. 2025 Feb 7;5(1):vbaf015. doi: 10.1093/bioadv/vbaf015. eCollection 2025. Bioinform Adv. 2025. PMID: 39968377 Free PMC article.
References
-
- World Health Organization. Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (2022). - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

