[Integrated analysis of serum untargeted metabolomics and targeted bile acid metabolomics for identification of diagnostic biomarkers for colorectal cancer]
- PMID: 37087590
- PMCID: PMC10122735
- DOI: 10.12122/j.issn.1673-4254.2023.03.15
[Integrated analysis of serum untargeted metabolomics and targeted bile acid metabolomics for identification of diagnostic biomarkers for colorectal cancer]
Abstract
Objective: To identify potential diagnostic biomarkers of colorectal cancer (CRC) using serum metabolomic technology for minimally invasive and efficient screening for CRC.
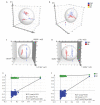
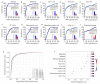
Methods: Serum samples from 79 healthy individuals and 82 CRC patients were analyzed by metabolomics using ultra-high-performance liquid chromatography-tandem highresolution mass spectrometry (UHPLC-HRMS). The differential metabolites between the two groups were analyzed using principal component analysis and orthogonal partial least squares discriminant analysis (OPLS-DA). Receiver operating characteristic curve (ROC) analysis was performed to identify the differential metabolites with good diagnostic performance (AUC>0.80) for CRC, and targeted bile acid metabolomics was used to verify the selected bile acids as biomarkers.
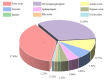
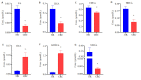
Results: Serum metabolic profiles differed significantly between the healthy individuals and CRC patients, and a total of 82 differential metabolites (mostly fatty acids and glycerophospholipids) were selected. ROC analysis identified 10 differential metabolites, including adenine, bilirubin, ACar 12:0, ACar 10:1, ACar 9:0, PC 18:2e, deoxycholic acid, chenodeoxycholic acid, ACar 14:1 and palmitoylcarnitine. One of these metabolites was significantly up-regulated and 9 were down-regulated in the serum of CRC patients (P < 0.05). Multivariate ROC analysis with support vector machine algorithm showed that the biomarker panel consisting of 7 differential metabolites had an AUC of 0.94 for CRC diagnosis. The results of targeted bile acid metabolomics were consistent with those of untargeted metabolomics. The serum levels of deoxycholic acid and chenodeoxycholic acid were significantly down-regulated in patients with CRC as compared with the healthy individuals (P < 0.05).
Conclusion: Metabolic disorders of fatty acids and glycerophospholipids are closely related wigh tumorigenesis of CRC. Ten differential metabolites show good performance for CRC diagnosis, and the panel consisting 7 of these metabolites has important diagnostic value for CRC. Deoxycholic acid and chenodeoxycholic acid may serve as potential diagnostic biomarkers of CRC.
目的: 应用血清代谢组学技术发现结直肠癌(CRC)的生物标志物以微创且高效地筛查CRC。
方法: 采用超高效液相色谱串联高分辨率质谱分析技术(UHPLC-HRMS)对79例健康对照(NR组)受试者和82例CRC患者(CRC组)的血清样本进行代谢组学分析。在两组血清代谢轮廓存在明显区别的基础上,采用单因素以及多元统计分析包括主成分分析和正交偏最小二乘判别分析进一步确定组间差异代谢物。以P < 0.05,倍数变化 < 0.67或>1.50,结合变量重要性投影>1.00为筛选标准发现差异代谢物,并通过受试者工作特征曲线分析(ROC)筛选出对CRC具有良好诊断效能(曲线下面积AUC>0.80)的潜在诊断标志物。同时,应用靶向胆汁酸代谢组学对所筛选的胆汁酸类肿瘤生物标志物进行靶向验证。
结果: NR组和CRC组的血清代谢谱存在明显差异。基于上述标准共筛选获得82种组间差异代谢物,以脂肪酸和甘油磷脂为主。ROC分析表明包括腺嘌呤、胆红素、ACar(乙酰肉碱)12:0、ACar 10:1、ACar 9:0、PC(磷脂酰胆碱)18:2e、去氧胆酸、鹅去氧胆酸、ACar 14:1、棕榈酰肉碱这10个差异代谢物其AUC值>0.80。其中,与NR组相比,1个差异代谢物在CRC组中呈现显著上调,其余9个差异代谢物均呈显著下调(P < 0.05)。此外,基于支持向量机算法的多变量ROC分析发现,其中7个差异代谢物组成的标志物组合的AUC值为0.94,对CRC具有出色的诊断效能。靶向胆汁酸代谢组学结果与非靶向代谢组学的结果一致,去氧胆酸和鹅去氧胆酸的相对血清水平较NR组在CRC中呈现显著下调趋势(P < 0.05)。
结论: 脂肪酸和甘油磷脂的代谢紊乱可能与CRC的形成密切相关。10个差异代谢物对CRC具有良好诊断效能,其中7个差异代谢物组成的标志物组合对CRC的临床诊断具有重要的应用潜力,特别是去氧胆酸和鹅去氧胆酸可作为CRC的潜在生物标志物。
Keywords: biomarkers; colorectal cancer; serum untargeted metabolomics; targeted bile acid metabolomics.
Figures


Similar articles
-
UHPLC-HRMS-based Multiomics to Explore the Potential Mechanisms and Biomarkers for Colorectal Cancer.BMC Cancer. 2024 May 27;24(1):644. doi: 10.1186/s12885-024-12321-7. BMC Cancer. 2024. PMID: 38802800 Free PMC article.
-
[Plasma metabolomics in a deep vein thrombosis rat model based on ultra-high performance liquid chromatography-electrostatic field orbitrap high resolution mass spectrometry].Se Pu. 2022 Aug;40(8):736-745. doi: 10.3724/SP.J.1123.2021.12024. Se Pu. 2022. PMID: 35903841 Free PMC article. Chinese.
-
Metabolomic Profiling Identified Serum Metabolite Biomarkers and Related Metabolic Pathways of Colorectal Cancer.Dis Markers. 2021 Dec 7;2021:6858809. doi: 10.1155/2021/6858809. eCollection 2021. Dis Markers. 2021. PMID: 34917201 Free PMC article. Clinical Trial.
-
Urine NMR Metabolomics for Precision Oncology in Colorectal Cancer.Int J Mol Sci. 2022 Sep 22;23(19):11171. doi: 10.3390/ijms231911171. Int J Mol Sci. 2022. PMID: 36232473 Free PMC article. Review.
-
Diagnostic performance of serum metabolites biomarker associated with colorectal adenoma: a systematic review.PeerJ. 2024 Sep 20;12:e18043. doi: 10.7717/peerj.18043. eCollection 2024. PeerJ. 2024. PMID: 39314843 Free PMC article.
Cited by
-
From multi-omics to predictive biomarker: AI in tumor microenvironment.Front Immunol. 2024 Dec 23;15:1514977. doi: 10.3389/fimmu.2024.1514977. eCollection 2024. Front Immunol. 2024. PMID: 39763649 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
