Transcription factor localization dynamics and DNA binding drive distinct promoter interpretations
- PMID: 37087734
- PMCID: PMC10292158
- DOI: 10.1016/j.celrep.2023.112426
Transcription factor localization dynamics and DNA binding drive distinct promoter interpretations
Abstract
Environmental information may be encoded in the temporal dynamics of transcription factor (TF) activation and subsequently decoded by gene promoters to enact stimulus-specific gene expression programs. Previous studies of this behavior focused on the encoding and decoding of information in TF nuclear localization dynamics, yet cells control the activity of TFs in myriad ways, including by regulating their ability to bind DNA. Here, we use light-controlled mutants of the yeast TF Msn2 as a model system to investigate how promoter decoding of TF localization dynamics is affected by changes in the ability of the TF to bind DNA. We find that yeast promoters directly decode the light-controlled localization dynamics of Msn2 and that the effects of changing Msn2 affinity on that decoding behavior are highly promoter dependent, illustrating how cells could regulate TF localization dynamics and DNA binding in concert for improved control of gene expression.
Keywords: CP: Molecular biology; gene expression; optogenetics; promoter decoding; signaling dynamics; transcription factors.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



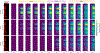

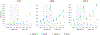
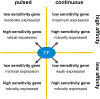
References
-
- Chen SY, Osimiri LC, Chevalier M, Bugaj LJ, Nguyen TH, Green-stein RA, Ng AH, Stewart-Ornstein J, Neves LT, and El-Samad H (2020). Optogenetic control reveals differential promoter interpretation of transcription factor nuclear translocation dynamics. Cell Syst. 11, 336–353.e24. 10.1016/j.cels.2020.08.009. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

